The genomic landscape of swine influenza A viruses in Southeast Asia
- PMID: 37552753
- PMCID: PMC10438389
- DOI: 10.1073/pnas.2301926120
The genomic landscape of swine influenza A viruses in Southeast Asia
Abstract
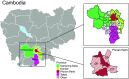
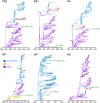
Swine are a primary source for the emergence of pandemic influenza A viruses. The intensification of swine production, along with global trade, has amplified the transmission and zoonotic risk of swine influenza A virus (swIAV). Effective surveillance is essential to uncover emerging virus strains; however gaps remain in our understanding of the swIAV genomic landscape in Southeast Asia. More than 4,000 nasal swabs were collected from pigs in Cambodia, yielding 72 IAV-positive samples by RT-qPCR and 45 genomic sequences. We unmasked the cocirculation of multiple lineages of genetically diverse swIAV of pandemic concern. Genomic analyses revealed a novel European avian-like H1N2 swIAV reassortant variant with North American triple reassortant internal genes, that emerged approximately seven years before its first detection in pigs in 2021. Using phylogeographic reconstruction, we identified south central China as the dominant source of swine viruses disseminated to other regions in China and Southeast Asia. We also identified nine distinct swIAV lineages in Cambodia, which diverged from their closest ancestors between two and 15 B.P., indicating significant undetected diversity in the region, including reverse zoonoses of human H1N1/2009 pandemic and H3N2 viruses. A similar period of cryptic circulation of swIAVs occurred in the decades before the H1N1/2009 pandemic. The hidden diversity of swIAV observed here further emphasizes the complex underlying evolutionary processes present in this region, reinforcing the importance of genomic surveillance at the human-swine interface for early warning of disease emergence to avoid future pandemics.
Keywords: European avian-like virus; evolution; pandemic; zoonotic.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

