Analysis of the complex between MBD2 and the histone deacetylase core of NuRD reveals key interactions critical for gene silencing
- PMID: 37552759
- PMCID: PMC10433457
- DOI: 10.1073/pnas.2307287120
Analysis of the complex between MBD2 and the histone deacetylase core of NuRD reveals key interactions critical for gene silencing
Abstract
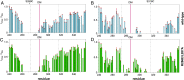
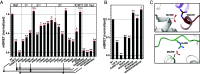
The nucleosome remodeling and deacetylase (NuRD) complex modifies nucleosome positioning and chromatin compaction to regulate gene expression. The methyl-CpG-binding domain proteins 2 and 3 (MBD2 and MBD3) play a critical role in complex formation; however, the molecular details of how they interact with other NuRD components have yet to be fully elucidated. We previously showed that an intrinsically disordered region (IDR) of MBD2 is necessary and sufficient to bind to the histone deacetylase core of NuRD. Building on that work, we have measured the inherent structural propensity of the MBD2-IDR using solvent and site-specific paramagnetic relaxation enhancement measurements. We then used the AlphaFold2 machine learning software to generate a model of the complex between MBD2 and the histone deacetylase core of NuRD. This model is remarkably consistent with our previous studies, including the current paramagnetic relaxation enhancement data. The latter suggests that the free MBD2-IDR samples conformations similar to the bound structure. We tested this model of the complex extensively by mutating key contact residues and measuring binding using an intracellular bioluminescent resonance energy transfer assay. Furthermore, we identified protein contacts that, when mutated, disrupted gene silencing by NuRD in a cell model of fetal hemoglobin regulation. Hence, this work provides insights into the formation of NuRD and highlights critical binding pockets that may be targeted to block gene silencing for therapy. Importantly, we show that AlphaFold2 can generate a credible model of a large complex that involves an IDR that folds upon binding.
Keywords: AlphaFold2; DNA methylation; MBD2-NuRD; chromatin remodeling; intrinsically disordered protein.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

