Peripheral macrophages drive CNS disease in the Ndufs4(-/-) model of Leigh syndrome
- PMID: 37552802
- PMCID: PMC10580015
- DOI: 10.1111/bpa.13192
Peripheral macrophages drive CNS disease in the Ndufs4(-/-) model of Leigh syndrome
Abstract
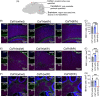
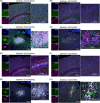
Subacute necrotizing encephalopathy, or Leigh syndrome (LS), is the most common pediatric presentation of genetic mitochondrial disease. LS is a multi-system disorder with severe neurologic, metabolic, and musculoskeletal symptoms. The presence of progressive, symmetric, and necrotizing lesions in the brainstem are a defining feature of the disease, and the major cause of morbidity and mortality, but the mechanisms underlying their pathogenesis have been elusive. Recently, we demonstrated that high-dose pexidartinib, a CSF1R inhibitor, prevents LS CNS lesions and systemic disease in the Ndufs4(-/-) mouse model of LS. While the dose-response in this study implicated peripheral immune cells, the immune populations involved have not yet been elucidated. Here, we used a targeted genetic tool, deletion of the colony-stimulating Factor 1 receptor (CSF1R) macrophage super-enhancer FIRE (Csf1rΔFIRE), to specifically deplete microglia and define the role of microglia in the pathogenesis of LS. Homozygosity for the Csf1rΔFIRE allele ablates microglia in both control and Ndufs4(-/-) animals, but onset of CNS lesions and sequalae in the Ndufs4(-/-), including mortality, are only marginally impacted by microglia depletion. The overall development of necrotizing CNS lesions is not altered, though microglia remain absent. Finally, histologic analysis of brainstem lesions provides direct evidence of a causal role for peripheral macrophages in the characteristic CNS lesions. These data demonstrate that peripheral macrophages play a key role in the pathogenesis of disease in the Ndufs4(-/-) model.
Keywords: CNS lesions; Leigh syndrome; microglia; mitochondrial disease; pediatric disease; subacute necrotizing encephalomyelopathy.
© 2023 The Authors. Brain Pathology published by John Wiley & Sons Ltd on behalf of International Society of Neuropathology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

