Spatial positioning of preimplantation mouse embryo cells is regulated by mTORC1 and m7G-cap-dependent translation at the 8- to 16-cell transition
- PMID: 37553074
- PMCID: PMC10409569
- DOI: 10.1098/rsob.230081
Spatial positioning of preimplantation mouse embryo cells is regulated by mTORC1 and m7G-cap-dependent translation at the 8- to 16-cell transition
Abstract
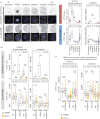
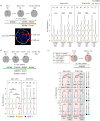
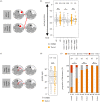
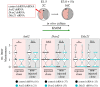
Preimplantation mouse embryo development involves temporal-spatial specification and segregation of three blastocyst cell lineages: trophectoderm, primitive endoderm and epiblast. Spatial separation of the outer-trophectoderm lineage from the two other inner-cell-mass (ICM) lineages starts with the 8- to 16-cell transition and concludes at the 32-cell stages. Accordingly, the ICM is derived from primary and secondary contributed cells; with debated relative EPI versus PrE potencies. We report generation of primary but not secondary ICM populations is highly dependent on temporal activation of mammalian target of Rapamycin (mTOR) during 8-cell stage M-phase entry, mediated via regulation of the 7-methylguanosine-cap (m7G-cap)-binding initiation complex (EIF4F) and linked to translation of mRNAs containing 5' UTR terminal oligopyrimidine (TOP-) sequence motifs, as knockdown of identified TOP-like motif transcripts impairs generation of primary ICM founders. However, mTOR inhibition-induced ICM cell number deficits in early blastocysts can be compensated by the late blastocyst stage, after inhibitor withdrawal; compensation likely initiated at the 32-cell stage when supernumerary outer cells exhibit molecular characteristics of inner cells. These data identify a novel mechanism specifically governing initial spatial segregation of mouse embryo blastomeres, that is distinct from those directing subsequent inner cell formation, contributing to germane segregation of late blastocyst lineages.
Keywords: EIF4EBP1/4EBP1; TOP-motif; cell fate; inner cell mass/ICM and cell positioning; mTOR/mTORC1; preimplantation mouse embryo.
Conflict of interest statement
All authors declare a complete lack of conflict of interest arising from the reported research.
Figures

Similar articles
-
Lineage segregation in human pre-implantation embryos is specified by YAP1 and TEAD1.Hum Reprod. 2023 Aug 1;38(8):1484-1498. doi: 10.1093/humrep/dead107. Hum Reprod. 2023. PMID: 37295962
-
Does mouse embryo primordial germ cell activation start before implantation as suggested by single-cell transcriptomics dynamics?Mol Hum Reprod. 2016 Mar;22(3):208-25. doi: 10.1093/molehr/gav072. Epub 2016 Jan 5. Mol Hum Reprod. 2016. PMID: 26740066
-
FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst.Development. 2010 Mar;137(5):715-24. doi: 10.1242/dev.043471. Development. 2010. PMID: 20147376
-
Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo.Genesis. 2013 Apr;51(4):219-33. doi: 10.1002/dvg.22368. Epub 2013 Feb 25. Genesis. 2013. PMID: 23349011 Free PMC article. Review.
-
Journey of the mouse primitive endoderm: from specification to maturation.Philos Trans R Soc Lond B Biol Sci. 2022 Dec 5;377(1865):20210252. doi: 10.1098/rstb.2021.0252. Epub 2022 Oct 17. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 36252215 Free PMC article. Review.
Cited by
-
Technology to the rescue: how to uncover the role of transposable elements in preimplantation development.Biochem Soc Trans. 2024 Jun 26;52(3):1349-1362. doi: 10.1042/BST20231262. Biochem Soc Trans. 2024. PMID: 38752836 Free PMC article. Review.
-
The translational oscillation in oocyte and early embryo development.Nucleic Acids Res. 2023 Dec 11;51(22):12076-12091. doi: 10.1093/nar/gkad996. Nucleic Acids Res. 2023. PMID: 37950888 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

