Differentiating brain metastasis from glioblastoma by time-dependent diffusion MRI
- PMID: 37553578
- PMCID: PMC10410879
- DOI: 10.1186/s40644-023-00595-2
Differentiating brain metastasis from glioblastoma by time-dependent diffusion MRI
Abstract
Background: This study was designed to investigate the use of time-dependent diffusion magnetic resonance imaging (MRI) parameters in distinguishing between glioblastomas and brain metastases.
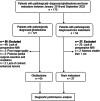
Methods: A retrospective study was conducted involving 65 patients with glioblastomas and 27 patients with metastases using a diffusion-weighted imaging sequence with oscillating gradient spin-echo (OGSE, 50 Hz) and a conventional pulsed gradient spin-echo (PGSE, 0 Hz) sequence. In addition to apparent diffusion coefficient (ADC) maps from two sequences (ADC50Hz and ADC0Hz), we generated maps of the ADC change (cADC): ADC50Hz - ADC0Hz and the relative ADC change (rcADC): (ADC50Hz - ADC0Hz)/ ADC0Hz × 100 (%).
Results: The mean and the fifth and 95th percentile values of each parameter in enhancing and peritumoral regions were compared between glioblastomas and metastases. The area under the receiver operating characteristic curve (AUC) values of the best discriminating indices were compared. In enhancing regions, none of the indices of ADC0Hz and ADC50Hz showed significant differences between metastases and glioblastomas. The mean cADC and rcADC values of metastases were significantly higher than those of glioblastomas (0.24 ± 0.12 × 10-3mm2/s vs. 0.14 ± 0.03 × 10-3mm2/s and 23.3 ± 9.4% vs. 14.0 ± 4.7%; all p < 0.01). In peritumoral regions, no significant difference in all ADC indices was observed between metastases and glioblastomas. The AUC values for the mean cADC (0.877) and rcADC (0.819) values in enhancing regions were significantly higher than those for ADC0Hz5th (0.595; all p < 0.001).
Conclusions: The time-dependent diffusion MRI parameters may be useful for differentiating brain metastases from glioblastomas.
Keywords: Diffusion; Glioblastoma; Magnetic resonance imaging; Neoplasm metastasis.
© 2023. International Cancer Imaging Society (ICIS).
Conflict of interest statement
Authors have no conflict of interest.
Figures






References
-
- Oh J, Cha S, Aiken AH, Han ET, Crane JC, Stainsby JA, et al. Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. J Magn Reson Imaging. 2005;21:701–8. 10.1002/jmri.20335. 10.1002/jmri.20335 - DOI - PubMed
-
- Eidel O, Neumann JO, Burth S, Kieslich PJ, Jungk C, Sahm F, et al. Automatic analysis of cellularity in glioblastoma and correlation with ADC Using trajectory analysis and automatic nuclei counting. PLoS One. 2016;11:e0160250. 10.1371/journal.pone.0160250. 10.1371/journal.pone.0160250 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

