Protein phosphatase 2A-B55β mediated mitochondrial p-GPX4 dephosphorylation promoted sorafenib-induced ferroptosis in hepatocellular carcinoma via regulating p53 retrograde signaling
- PMID: 37554285
- PMCID: PMC10405852
- DOI: 10.7150/thno.82132
Protein phosphatase 2A-B55β mediated mitochondrial p-GPX4 dephosphorylation promoted sorafenib-induced ferroptosis in hepatocellular carcinoma via regulating p53 retrograde signaling
Abstract
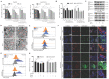
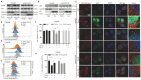
Rationale: As a key endogenous negative regulator of ferroptosis, glutathione peroxidase 4 (GPX4) can regulate its antioxidant function through multiple post-translational modification pathways. However, the effects of the phosphorylation/dephosphorylation status of GPX4 on the regulation of inducible ferroptosis in hepatocellular carcinoma (HCC) remain unclear. Methods: To investigate the effects and molecular mechanism of GPX4 phosphorylation/dephosphorylation modification on ferroptosis in HCC cells. Sorafenib (Sora) was used to establish the ferroptosis model in HCC cells in vitro. Using the site-directed mutagenesis method, we generated the mimic GPX4 phosphorylation or dephosphorylation HCC cell lines at specific serine sites of GPX4. The effects of GPX4 phosphorylation/dephosphorylation modification on ferroptosis in HCC cells were examined. The interrelationships among GPX4, p53, and protein phosphatase 2A-B55β subunit (PP2A-B55β) were also explored. To explore the synergistic anti-tumor effects of PP2A activation on Sora-administered HCC, we established PP2A-B55β overexpression xenograft tumors in a nude mice model in vivo. Results: In the Sora-induced ferroptosis model of HCC in vitro, decreased levels of cytoplasmic and mitochondrial GPX4, mitochondrial dysfunction, and enhanced p53 retrograde signaling occurred under Sora treatment. Further, we found that mitochondrial p53 retrograded remarkably into the nucleus and aggravated Sora-induced ferroptosis. The phosphorylation status of GPX4 at the serine 2 site (GPX4Ser2) revealed that mitochondrial p-GPX4Ser2 dephosphorylation was positively associated with ferroptosis, and the mechanism might be related to mitochondrial p53 retrograding into the nucleus. In HCC cells overexpressing PP2A-B55β, it was found that PP2A-B55β directly interacted with mitochondrial GPX4 and promoted Sora-induced ferroptosis in HCC. Further, PP2A-B55β reduced the interaction between mitochondrial GPX4 and p53, leading to mitochondrial p53 retrograding into the nucleus. Moreover, it was confirmed that PP2A-B55β enhanced the ferroptosis-mediated tumor growth inhibition and mitochondrial p53 retrograde signaling in the Sora-treated HCC xenograft tumors. Conclusion: Our data uncovered that the PP2A-B55β/p-GPX4Ser2/p53 axis was a novel regulatory pathway of Sora-induced ferroptosis. Mitochondrial p-GPX4Ser2 dephosphorylation triggered ferroptosis via inducing mitochondrial p53 retrograding into the nucleus, and PP2A-B55β was an upstream signal modulator responsible for mitochondrial p-GPX4Ser2 dephosphorylation. Our findings might serve as a potential theranostic strategy to enhance the efficacy of Sora in HCC treatment through the targeted intervention of p-GPX4 dephosphorylation via PP2A-B55β activation.
Keywords: Ferroptosis; Glutathione peroxidase 4; Hepatocellular carcinoma; Protein phosphatase 2A-B55β subunit; Retrograding p53 signal.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. - PubMed
-
- Chen J, Jin R, Zhao J, Liu J, Ying H, Yan H. et al. Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett. 2015;367:1–11. - PubMed
-
- Louandre C, Marcq I, Bouhlal H, Lachaier E, Godin C, Saidak Z. et al. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015;356:971–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

