Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases
- PMID: 37554450
- PMCID: PMC10404741
- DOI: 10.1016/j.isci.2023.107403
Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases
Abstract
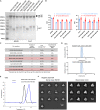
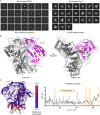
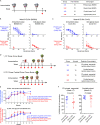
Soluble HIV-1-envelope (Env) trimers elicit immune responses that target their solvent-exposed protein bases, the result of removing these trimers from their native membrane-bound context. To assess whether glycosylation could limit these base responses, we introduced sequons encoding potential N-linked glycosylation sites (PNGSs) into base-proximal regions. Expression and antigenic analyses indicated trimers bearing six-introduced PNGSs to have reduced base recognition. Cryo-EM analysis revealed trimers with introduced PNGSs to be prone to disassembly and introduced PNGS to be disordered. Protein-base and glycan-base trimers induced reciprocally symmetric ELISA responses, in which only a small fraction of the antibody response to glycan-base trimers recognized protein-base trimers and vice versa. EM polyclonal epitope mapping revealed glycan-base trimers -even those that were stable biochemically- to elicit antibodies that recognized disassembled trimers. Introduced glycans can thus mask the protein base but their introduction may yield neo-epitopes that dominate the immune response.
Keywords: Molecular structure; Virology.
Conflict of interest statement
NIH has submitted a PCG patent application (PCT/US2023/065009) for Env trimers described in this manuscript on which A.S.O., C.C., T.Z., D.H.R., A.C., R.R., Y.Y., and P.D.K. are co-inventors. The other authors declare no competing interests.
Figures





References
-
- Kwon Y.D., Pancera M., Acharya P., Georgiev I.S., Crooks E.T., Gorman J., Joyce M.G., Guttman M., Ma X., Narpala S., et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

