De novo genome assembly and comparative genomics for the colonial ascidian Botrylloides violaceus
- PMID: 37555394
- PMCID: PMC10542563
- DOI: 10.1093/g3journal/jkad181
De novo genome assembly and comparative genomics for the colonial ascidian Botrylloides violaceus
Abstract
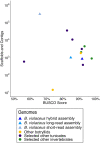
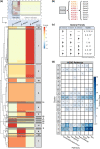
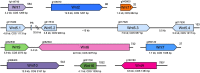
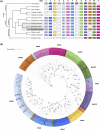
Ascidians have the potential to reveal fundamental biological insights related to coloniality, regeneration, immune function, and the evolution of these traits. This study implements a hybrid assembly technique to produce a genome assembly and annotation for the botryllid ascidian, Botrylloides violaceus. A hybrid genome assembly was produced using Illumina, Inc. short and Oxford Nanopore Technologies long-read sequencing technologies. The resulting assembly is comprised of 831 contigs, has a total length of 121 Mbp, N50 of 1 Mbp, and a BUSCO score of 96.1%. Genome annotation identified 13 K protein-coding genes. Comparative genomic analysis with other tunicates reveals patterns of conservation and divergence within orthologous gene families even among closely related species. Characterization of the Wnt gene family, encoding signaling ligands involved in development and regeneration, reveals conserved patterns of subfamily presence and gene copy number among botryllids. This supports the use of genomic data from nonmodel organisms in the investigation of biological phenomena.
Keywords: Illumina; Oxford Nanopore; Wnt; comparative genomics; genome assembly; tunicate; Botrylloides violaceus.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
