Experimental validation of the mechanism of stomatal development diversification
- PMID: 37555400
- PMCID: PMC10540739
- DOI: 10.1093/jxb/erad279
Experimental validation of the mechanism of stomatal development diversification
Abstract
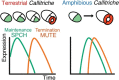
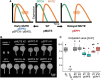
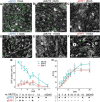
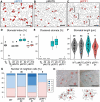
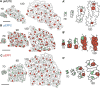
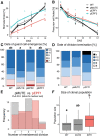
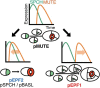
Stomata are the structures responsible for gas exchange in plants. The established framework for stomatal development is based on the model plant Arabidopsis, but diverse patterns of stomatal development have been observed in other plant lineages and species. The molecular mechanisms behind these diversified patterns are still poorly understood. We recently proposed a model for the molecular mechanisms of the diversification of stomatal development based on the genus Callitriche (Plantaginaceae), according to which a temporal shift in the expression of key stomatal transcription factors SPEECHLESS and MUTE leads to changes in the behavior of meristemoids (stomatal precursor cells). In the present study, we genetically manipulated Arabidopsis to test this model. By altering the timing of MUTE expression, we successfully generated Arabidopsis plants with early differentiation or prolonged divisions of meristemoids, as predicted by the model. The epidermal morphology of the generated lines resembled that of species with prolonged or no meristemoid divisions. Thus, the evolutionary process can be reproduced by varying the SPEECHLESS to MUTE transition. We also observed unexpected phenotypes, which indicated the participation of additional factors in the evolution of the patterns observed in nature. This study provides novel experimental insights into the diversification of meristemoid behaviors.
Keywords: Callitriche; Amplifying division; Arabidopsis; MUTE; SPEECHLESS; evo-devo; guard cells; meristemoid; stomata; stomatal development.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bergmann DC, Lukowitz W, Somerville CR.. 2004. Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

