Comparison of SARS-CoV-2 entry inhibitors based on ACE2 receptor or engineered Spike-binding peptides
- PMID: 37555663
- PMCID: PMC10506483
- DOI: 10.1128/jvi.00684-23
Comparison of SARS-CoV-2 entry inhibitors based on ACE2 receptor or engineered Spike-binding peptides
Abstract
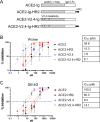
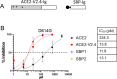
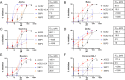
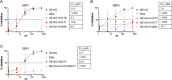
With increasing resistance of SARS-CoV-2 variants to antibodies, there is interest in developing entry inhibitors that target essential receptor-binding regions of the viral Spike protein and thereby present a high bar for viral resistance. Such inhibitors could be derivatives of the viral receptor, ACE2, or peptides engineered to interact specifically with the Spike receptor-binding pocket. We compared the efficacy of a series of both types of entry inhibitors, constructed as fusions to an antibody Fc domain. Such a design can increase protein stability and act to both neutralize free virus and recruit effector functions to clear infected cells. We tested the reagents against prototype variants of SARS-CoV-2, using both Spike pseudotyped vesicular stomatitis virus vectors and replication-competent viruses. These analyses revealed that an optimized ACE2 derivative could neutralize all variants we tested with high efficacy. In contrast, the Spike-binding peptides had varying activities against different variants, with resistance observed in the Spike proteins from Beta, Gamma, and Omicron (BA.1 and BA.5). The resistance mapped to mutations at Spike residues K417 and N501 and could be overcome for one of the peptides by linking two copies in tandem, effectively creating a tetrameric reagent in the Fc fusion. Finally, both the optimized ACE2 and tetrameric peptide inhibitors provided some protection to human ACE2 transgenic mice challenged with the SARS-CoV-2 Delta variant, which typically causes death in this model within 7-9 days. IMPORTANCE The increasing resistance of SARS-CoV-2 variants to therapeutic antibodies has highlighted the need for new treatment options, especially in individuals who do not respond to vaccination. Receptor decoys that block viral entry are an attractive approach because of the presumed high bar to developing viral resistance. Here, we compare two entry inhibitors based on derivatives of the ACE2 receptor, or engineered peptides that bind to the receptor-binding pocket of the SARS-CoV-2 Spike protein. In each case, the inhibitors were fused to immunoglobulin Fc domains, which can further enhance therapeutic properties, and compared for activity against different SARS-CoV-2 variants. Potent inhibition against multiple SARS-CoV-2 variants was demonstrated in vitro, and even relatively low single doses of optimized reagents provided some protection in a mouse model, confirming their potential as an alternative to antibody therapies.
Keywords: ACE2; SARS-CoV-2; Spike; entry inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation.Chem Biol Interact. 2022 Dec 1;368:110244. doi: 10.1016/j.cbi.2022.110244. Epub 2022 Nov 3. Chem Biol Interact. 2022. PMID: 36336003 Free PMC article.
-
Characterization of SARS-CoV-2 Variants B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.618 by Cell Entry and Immune Evasion.mBio. 2022 Apr 26;13(2):e0009922. doi: 10.1128/mbio.00099-22. Epub 2022 Mar 10. mBio. 2022. PMID: 35266815 Free PMC article.
-
SARS-CoV-2 strains bearing Omicron BA.1 spike replicate in C57BL/6 mice.Front Immunol. 2024 Apr 29;15:1383612. doi: 10.3389/fimmu.2024.1383612. eCollection 2024. Front Immunol. 2024. PMID: 38742107 Free PMC article.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
-
David versus goliath: ACE2-Fc receptor traps as potential SARS-CoV-2 inhibitors.MAbs. 2022 Jan-Dec;14(1):2057832. doi: 10.1080/19420862.2022.2057832. MAbs. 2022. PMID: 35380919 Free PMC article. Review.
Cited by
-
SCARF Genes in COVID-19 and Kidney Disease: A Path to Comorbidity-Specific Therapies.Int J Mol Sci. 2023 Nov 8;24(22):16078. doi: 10.3390/ijms242216078. Int J Mol Sci. 2023. PMID: 38003268 Free PMC article. Review.
-
FeLIX is a restriction factor for mammalian retrovirus infection.J Virol. 2024 Apr 16;98(4):e0177123. doi: 10.1128/jvi.01771-23. Epub 2024 Mar 5. J Virol. 2024. PMID: 38440982 Free PMC article.
-
Susceptibility and Resistance of SARS-CoV-2 Variants to LCB1 and Its Multivalent Derivatives.Viruses. 2023 Dec 25;16(1):36. doi: 10.3390/v16010036. Viruses. 2023. PMID: 38257736 Free PMC article.
-
Reprogramming human B cells with custom heavy-chain antibodies.Nat Biomed Eng. 2024 Dec;8(12):1700-1714. doi: 10.1038/s41551-024-01240-4. Epub 2024 Jul 22. Nat Biomed Eng. 2024. PMID: 39039240
References
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. Addendum: a pneumonia outbreak associated with a new Coronavirus of probable bat origin. Nature 588:E6. doi:10.1038/s41586-020-2951-z - DOI - PMC - PubMed
-
- Johns Hopkins University Coronavirus Resource Center. Baltimore MD.
-
- CDC COVID data Tracker. https://covid.cdc.gov/covid-data-tracker/#variant-proportions.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

