iCpG-Pos: an accurate computational approach for identification of CpG sites using positional features on single-cell whole genome sequence data
- PMID: 37555812
- PMCID: PMC10444964
- DOI: 10.1093/bioinformatics/btad474
iCpG-Pos: an accurate computational approach for identification of CpG sites using positional features on single-cell whole genome sequence data
Abstract
Motivation: The investigation of DNA methylation can shed light on the processes underlying human well-being and help determine overall human health. However, insufficient coverage makes it challenging to implement single-stranded DNA methylation sequencing technologies, highlighting the need for an efficient prediction model. Models are required to create an understanding of the underlying biological systems and to project single-cell (methylated) data accurately.
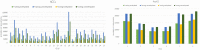
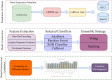
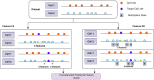
Results: In this study, we developed positional features for predicting CpG sites. Positional characteristics of the sequence are derived using data from CpG regions and the separation between nearby CpG sites. Multiple optimized classifiers and different ensemble learning approaches are evaluated. The OPTUNA framework is used to optimize the algorithms. The CatBoost algorithm followed by the stacking algorithm outperformed existing DNA methylation identifiers.
Availability and implementation: The data and methodologies used in this study are openly accessible to the research community. Researchers can access the positional features and algorithms used for predicting CpG site methylation patterns. To achieve superior performance, we employed the CatBoost algorithm followed by the stacking algorithm, which outperformed existing DNA methylation identifiers. The proposed iCpG-Pos approach utilizes only positional features, resulting in a substantial reduction in computational complexity compared to other known approaches for detecting CpG site methylation patterns. In conclusion, our study introduces a novel approach, iCpG-Pos, for predicting CpG site methylation patterns. By focusing on positional features, our model offers both accuracy and efficiency, making it a promising tool for advancing DNA methylation research and its applications in human health and well-being.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
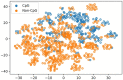
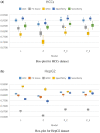
References
-
- Akiba T, Sano S, Yanase T et al. Optuna: A next-generation hyperparameter optimization framework. In: Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage AK, USA, August 4–8, 2019. New York, NY, United States: Association for Computing Machinery, 2019, 2623–31.
-
- Bhasin M, Zhang H, Reinherz EL et al. Prediction of methylated CpGs in DNA sequences using a support vector machine. FEBS Lett 2005;579:4302–8. - PubMed
-
- Chou K-C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 2001;43:246–55. - PubMed

