Rejection of inappropriate synaptic partners in mouse retina mediated by transcellular FLRT2-UNC5 signaling
- PMID: 37557174
- PMCID: PMC10615732
- DOI: 10.1016/j.devcel.2023.07.011
Rejection of inappropriate synaptic partners in mouse retina mediated by transcellular FLRT2-UNC5 signaling
Abstract
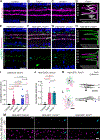
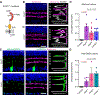
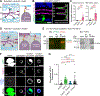
During nervous system development, neurons choose synaptic partners with remarkable specificity; however, the cell-cell recognition mechanisms governing rejection of inappropriate partners remain enigmatic. Here, we show that mouse retinal neurons avoid inappropriate partners by using the FLRT2-uncoordinated-5 (UNC5) receptor-ligand system. Within the inner plexiform layer (IPL), FLRT2 is expressed by direction-selective (DS) circuit neurons, whereas UNC5C/D are expressed by non-DS neurons projecting to adjacent IPL sublayers. In vivo gain- and loss-of-function experiments demonstrate that FLRT2-UNC5 binding eliminates growing DS dendrites that have strayed from the DS circuit IPL sublayers. Abrogation of FLRT2-UNC5 binding allows mistargeted arbors to persist, elaborate, and acquire synapses from inappropriate partners. Conversely, UNC5C misexpression within DS circuit sublayers inhibits dendrite growth and drives arbors into adjacent sublayers. Mechanistically, UNC5s promote dendrite elimination by interfering with FLRT2-mediated adhesion. Based on their broad expression, FLRT-UNC5 recognition is poised to exert widespread effects upon synaptic partner choices across the nervous system.
Keywords: dendrite; latrophilin; retina; retinal ganglion cell; starburst amacrine cell; synaptogenesis.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

