Modifications of lipid pathways restrict SARS-CoV-2 propagation in human induced pluripotent stem cell-derived 3D airway organoids
- PMID: 37557954
- PMCID: PMC11156708
- DOI: 10.1016/j.jare.2023.08.005
Modifications of lipid pathways restrict SARS-CoV-2 propagation in human induced pluripotent stem cell-derived 3D airway organoids
Abstract
Background: Modifications of lipid metabolism were closely associated with the manifestations and prognosis of coronavirus disease of 2019 (COVID-19). Pre-existing metabolic conditions exacerbated the severity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection while modulations of aberrant lipid metabolisms alleviated the manifestations. To elucidate the underlying mechanisms, an experimental platform that reproduces human respiratory physiology is required.
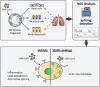
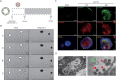
Methods: Here we generated induced pluripotent stem cell-derived airway organoids (iPSC-AOs) that resemble the human native airway. Single-cell sequencing (ScRNAseq) and microscopic examination verified the cellular heterogeneity and microstructures of iPSC-AOs, respectively. We subjected iPSC-AOs to SARS-CoV-2 infection and investigated the treatment effect of lipid modifiers statin drugs on viral pathogenesis, gene expression, and the intracellular trafficking of the SARS-CoV-2 entry receptor angiotensin-converting enzyme-2 (ACE-2).
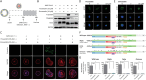
Results: In SARS-CoV-2-infected iPSC-AOs, immunofluorescence staining detected the SARS-CoV-2 spike (S) and nucleocapsid (N) proteins and bioinformatics analysis further showed the aberrant enrichment of lipid-associated pathways. In addition, SARS-CoV-2 hijacked the host RNA replication machinery and generated the new isoforms of a high-density lipoprotein constituent apolipoprotein A1 (APOA1) and the virus-scavenging protein deleted in malignant brain tumors 1 (DMBT1). Manipulating lipid homeostasis using cholesterol-lowering drugs (e.g. Statins) relocated the viral entry receptor angiotensin-converting enzyme-2 (ACE-2) and decreased N protein expression, leading to the reduction of SARS-CoV-2 entry and replication. The same lipid modifications suppressed the entry of luciferase-expressing SARS-CoV-2 pseudoviruses containing the S proteins derived from different SARS-CoV-2 variants, i.e. wild-type, alpha, delta, and omicron.
Conclusions: Together, our data demonstrated that modifications of lipid pathways restrict SARS-CoV-2 propagation in the iPSC-AOs, which the inhibition is speculated through the translocation of ACE2 from the cell membrane to the cytosol. Considering the highly frequent mutation and generation of SARS-CoV-2 variants, targeting host metabolisms of cholesterol or other lipids may represent an alternative approach against SARS-CoV-2 infection.
Keywords: Airway organoid; Angiotensin-converting enzyme 2; Induced pluripotent stem cell; Severe acute respiratory syndrome coronavirus 2; Single cell RNA-sequencing.
Copyright © 2024. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. - PubMed
-
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

