Machine learning models of plasma proteomic data predict mood in chronic stroke and tie it to aberrant peripheral immune responses
- PMID: 37557961
- PMCID: PMC10792657
- DOI: 10.1016/j.bbi.2023.08.002
Machine learning models of plasma proteomic data predict mood in chronic stroke and tie it to aberrant peripheral immune responses
Abstract
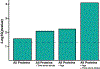
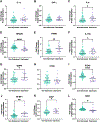
Post-stroke depression is common, long-lasting and associated with severe morbidity and death, but mechanisms are not well-understood. We used a broad proteomics panel and developed a machine learning algorithm to determine whether plasma protein data can predict mood in people with chronic stroke, and to identify proteins and pathways associated with mood. We used Olink to measure 1,196 plasma proteins in 85 participants aged 25 and older who were between 5 months and 9 years after ischemic stroke. Mood was assessed with the Stroke Impact Scale mood questionnaire (SIS3). Machine learning multivariable regression models were constructed to estimate SIS3 using proteomics data, age, and time since stroke. We also dichotomized participants into better mood (SIS3 > 63) or worse mood (SIS3 ≤ 63) and analyzed candidate proteins. Machine learning models verified that there is indeed a relationship between plasma proteomic data and mood in chronic stroke, with the most accurate prediction of mood occurring when we add age and time since stroke. At the individual protein level, no single protein or set of proteins predicts mood. But by using univariate analyses of the proteins most highly associated with mood we produced a model of chronic post-stroke depression. We utilized the fact that this list contained many proteins that are also implicated in major depression. Also, over 80% of immune proteins that correlate with mood were higher with worse mood, implicating a broadly overactive immune system in chronic post-stroke depression. Finally, we used a comprehensive literature review of major depression and acute post-stroke depression. We propose that in chronic post-stroke depression there is over-activation of the immune response that then triggers changes in serotonin activity and neuronal plasticity leading to depressed mood.
Keywords: Biomarker; Depression; Machine learning; Proteomics; Proximity extension assay; Stroke.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



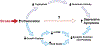
References
-
- Virani SS, Alonso A, Benjamin EJ, et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation. 2020;141(9):e139–e596. - PubMed
-
- Ferrari F, Villa RF. The Neurobiology of Depression: an Integrated Overview from Biological Theories to Clinical Evidence. Mol Neurobiol. 2017;54(7):4847–4865. - PubMed
-
- Ayerbe L, Ayis S, Wolfe CDA, Rudd AG. Natural history, predictors and outcomes of depression after stroke: systematic review and meta-analysis. Br J Psychiatry. 2013;202(1):14–21. - PubMed
-
- Towfighi A, Ovbiagele B, El Husseini N, et al. Poststroke Depression: A Scientific Statement for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke. 2017;48(2):e30–e43. - PubMed
-
- Richards D. Prevalence and clinical course of depression: a review. Clin Psychol Rev. 2011;31(7):1117–1125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

