Use of genome sequencing to hunt for cryptic second-hit variants: analysis of 31 cases recruited to the 100 000 Genomes Project
- PMID: 37558402
- PMCID: PMC10715503
- DOI: 10.1136/jmg-2023-109362
Use of genome sequencing to hunt for cryptic second-hit variants: analysis of 31 cases recruited to the 100 000 Genomes Project
Abstract
Background: Current clinical testing methods used to uncover the genetic basis of rare disease have inherent limitations, which can lead to causative pathogenic variants being missed. Within the rare disease arm of the 100 000 Genomes Project (100kGP), families were recruited under the clinical indication 'single autosomal recessive mutation in rare disease'. These participants presented with strong clinical suspicion for a specific autosomal recessive disorder, but only one suspected pathogenic variant had been identified through standard-of-care testing. Whole genome sequencing (WGS) aimed to identify cryptic 'second-hit' variants.
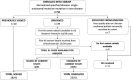
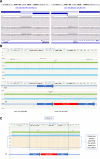
Methods: To investigate the 31 families with available data that remained unsolved following formal review within the 100kGP, SVRare was used to aggregate structural variants present in <1% of 100kGP participants. Small variants were assessed using population allele frequency data and SpliceAI. Literature searches and publicly available online tools were used for further annotation of pathogenicity.
Results: Using these strategies, 8/31 cases were solved, increasing the overall diagnostic yield of this cohort from 10/41 (24.4%) to 18/41 (43.9%). Exemplar cases include a patient with cystic fibrosis harbouring a novel exonic LINE1 insertion in CFTR and a patient with generalised arterial calcification of infancy with complex interlinked duplications involving exons 2-6 of ENPP1. Although ambiguous by short-read WGS, the ENPP1 variant structure was resolved using optical genome mapping and RNA analysis.
Conclusion: Systematic examination of cryptic variants across a multi-disease cohort successfully identifies additional pathogenic variants. WGS data analysis in autosomal recessive rare disease should consider complex structural and small intronic variants as potentially pathogenic second hits.
Keywords: diagnosis; genetic diseases, inborn; genetics, medical; genomics; sequence analysis, DNA.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


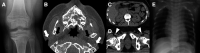
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
