A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis
- PMID: 37558666
- PMCID: PMC10412576
- DOI: 10.1038/s41467-023-40541-1
A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis
Erratum in
-
Author Correction: A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis.Nat Commun. 2023 Nov 27;14(1):7751. doi: 10.1038/s41467-023-43699-w. Nat Commun. 2023. PMID: 38012189 Free PMC article. No abstract available.
-
Author Correction: A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis.Nat Commun. 2024 Mar 26;15(1):2658. doi: 10.1038/s41467-024-47041-w. Nat Commun. 2024. PMID: 38531897 Free PMC article. No abstract available.
Abstract
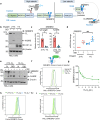
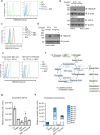
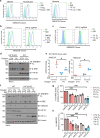
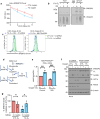
Cholesterol biosynthesis is a highly regulated, oxygen-dependent pathway, vital for cell membrane integrity and growth. In fungi, the dependency on oxygen for sterol production has resulted in a shared transcriptional response, resembling prolyl hydroxylation of Hypoxia Inducible Factors (HIFs) in metazoans. Whether an analogous metazoan pathway exists is unknown. Here, we identify Sterol Regulatory Element Binding Protein 2 (SREBP2), the key transcription factor driving sterol production in mammals, as an oxygen-sensitive regulator of cholesterol synthesis. SREBP2 degradation in hypoxia overrides the normal sterol-sensing response, and is HIF independent. We identify MARCHF6, through its NADPH-mediated activation in hypoxia, as the main ubiquitin ligase controlling SREBP2 stability. Hypoxia-mediated degradation of SREBP2 protects cells from statin-induced cell death by forcing cells to rely on exogenous cholesterol uptake, explaining why many solid organ tumours become auxotrophic for cholesterol. Our findings therefore uncover an oxygen-sensitive pathway for governing cholesterol synthesis through regulated SREBP2-dependent protein degradation.
© 2023. Springer Nature Limited.
Conflict of interest statement
J.A.N. receives a Pfizer ITEN discovery grant for unrelated work to this manuscript. Other authors declare no competing interests.
Figures


Similar articles
-
FoxO4 interacts with the sterol regulatory factor SREBP2 and the hypoxia inducible factor HIF2α at the CYP51 promoter.J Lipid Res. 2014 Mar;55(3):431-42. doi: 10.1194/jlr.M043521. Epub 2013 Dec 18. J Lipid Res. 2014. PMID: 24353279 Free PMC article.
-
Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism.J Biol Chem. 2004 Nov 19;279(47):48801-7. doi: 10.1074/jbc.M407817200. Epub 2004 Sep 8. J Biol Chem. 2004. PMID: 15358760
-
Inflammatory stress signaling via NF-kB alters accessible cholesterol to upregulate SREBP2 transcriptional activity in endothelial cells.Elife. 2022 Aug 12;11:e79529. doi: 10.7554/eLife.79529. Elife. 2022. PMID: 35959888 Free PMC article.
-
A paREDOX in the control of cholesterol biosynthesis: Does the NADPH sensor and E3 ubiquitin ligase MARCHF6 protect mammalian cells during oxidative stress by controlling sterol biosynthesis?Bioessays. 2024 Jul;46(7):e2400073. doi: 10.1002/bies.202400073. Epub 2024 May 17. Bioessays. 2024. PMID: 38760877 Review.
-
Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing.IUBMB Life. 2001 Jul;52(1-2):43-7. doi: 10.1080/15216540252774757. IUBMB Life. 2001. PMID: 11795592 Review.
Cited by
-
A tale of 2 gasses, 1 regulator, and cholesterol homeostasis.PLoS Biol. 2023 Nov 22;21(11):e3002401. doi: 10.1371/journal.pbio.3002401. eCollection 2023 Nov. PLoS Biol. 2023. PMID: 37992072 Free PMC article.
-
Biophysical interplay between extracellular matrix remodeling and hypoxia signaling in regulating cancer metastasis.Front Cell Dev Biol. 2024 Mar 13;12:1335636. doi: 10.3389/fcell.2024.1335636. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38544822 Free PMC article. Review.
-
Hypoxia exposure impairs male fertility via inhibiting Septin2-mediated spermatogonial proliferation.Hum Reprod Open. 2025 May 14;2025(3):hoaf027. doi: 10.1093/hropen/hoaf027. eCollection 2025. Hum Reprod Open. 2025. PMID: 40487848 Free PMC article.
-
March6 Protects Against Acute Kidney Injury by Suppressing Renal Tubular Epithelial Cell Ferroptosis Through the Destabilization of P53 and ACSL4 Proteins.Inflammation. 2025 May 30. doi: 10.1007/s10753-025-02319-z. Online ahead of print. Inflammation. 2025. PMID: 40445517
-
Intertwined regulators: hypoxia pathway proteins, microRNAs, and phosphodiesterases in the control of steroidogenesis.Pflugers Arch. 2024 Sep;476(9):1383-1398. doi: 10.1007/s00424-024-02921-4. Epub 2024 Feb 15. Pflugers Arch. 2024. PMID: 38355819 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

