Auranofin targets UBA1 and enhances UBA1 activity by facilitating ubiquitin trans-thioesterification to E2 ubiquitin-conjugating enzymes
- PMID: 37558718
- PMCID: PMC10412574
- DOI: 10.1038/s41467-023-40537-x
Auranofin targets UBA1 and enhances UBA1 activity by facilitating ubiquitin trans-thioesterification to E2 ubiquitin-conjugating enzymes
Abstract
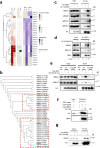
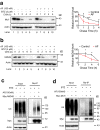
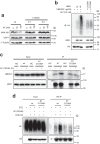
UBA1 is the primary E1 ubiquitin-activating enzyme responsible for generation of activated ubiquitin required for ubiquitination, a process that regulates stability and function of numerous proteins. Decreased or insufficient ubiquitination can cause or drive aging and many diseases. Therefore, a small-molecule enhancing UBA1 activity could have broad therapeutic potential. Here we report that auranofin, a drug approved for the treatment of rheumatoid arthritis, is a potent UBA1 activity enhancer. Auranofin binds to the UBA1's ubiquitin fold domain and conjugates to Cys1039 residue. The binding enhances UBA1 interactions with at least 20 different E2 ubiquitin-conjugating enzymes, facilitating ubiquitin charging to E2 and increasing the activities of seven representative E3s in vitro. Auranofin promotes ubiquitination and degradation of misfolded ER proteins during ER-associated degradation in cells at low nanomolar concentrations. It also facilitates outer mitochondrial membrane-associated degradation. These findings suggest that auranofin can serve as a much-needed tool for UBA1 research and therapeutic exploration.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

