An analysis of codon utilization patterns in the chloroplast genomes of three species of Coffea
- PMID: 37558997
- PMCID: PMC10413492
- DOI: 10.1186/s12863-023-01143-4
An analysis of codon utilization patterns in the chloroplast genomes of three species of Coffea
Abstract
Background: The chloroplast genome of plants is known for its small size and low mutation and recombination rates, making it a valuable tool in plant phylogeny, molecular evolution, and population genetics studies. Codon usage bias, an important evolutionary feature, provides insights into species evolution, gene function, and the expression of exogenous genes. Coffee, a key crop in the global tropical agricultural economy, trade, and daily life, warrants investigation into its codon usage bias to guide future research, including the selection of efficient heterologous expression systems for coffee genetic transformation.
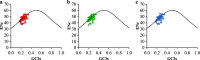
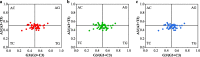
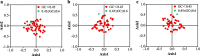
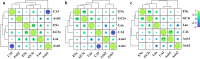
Results: Analysis of the codon utilization patterns in the chloroplast genomes of three Coffea species revealed a high degree of similarity among them. All three species exhibited similar base compositions, with high A/T content and low G/C content and a preference for A/T-ending codons. Among the 30 high-frequency codons identified, 96.67% had A/T endings. Fourteen codons were identified as ideal. Multiple mechanisms, including natural selection, were found to influence the codon usage patterns in the three coffee species, as indicated by ENc-GC3s mapping, PR2 analysis, and neutral analysis. Nicotiana tabacum and Saccharomyces cerevisiae have potential value as the heterologous expression host for three species of coffee genes.
Conclusion: This study highlights the remarkable similarity in codon usage patterns among the three coffee genomes, primarily driven by natural selection. Understanding the gene expression characteristics of coffee and elucidating the laws governing its genetic evolution are facilitated by investigating the codon preferences in these species. The findings can enhance the efficacy of exogenous gene expression and serve as a basis for future studies on coffee evolution.
Keywords: Chloroplast genome; Codon usage bias; Coffee; Heterologous expression.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Narko T, Wibowo MS, Damayanti S, Wibowo I. Acute Toxicity Tests of Fermented Robusta Green Coffee Using Zebrafish Embryos (Danio rerio) Pharmacognosy J. 2020;12(3):485–492.
-
- Homan DJ, Mobarhan S. Coffee: good, bad, or just fun? A critical review of coffee's effects on liver enzymes. Nutr Rev. 2006;64(1):43–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
