Identification of a Protein Arginine Methyltransferase 7 (PRMT7)/Protein Arginine Methyltransferase 9 (PRMT9) Inhibitor
- PMID: 37560786
- PMCID: PMC10578352
- DOI: 10.1021/acs.jmedchem.3c01030
Identification of a Protein Arginine Methyltransferase 7 (PRMT7)/Protein Arginine Methyltransferase 9 (PRMT9) Inhibitor
Abstract
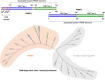
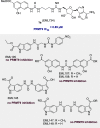
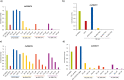
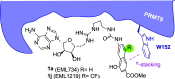
Less studied than the other protein arginine methyltransferase isoforms, PRMT7 and PRMT9 have recently been identified as important therapeutic targets. Yet, most of their biological roles and functions are still to be defined, as well as the structural requirements that could drive the identification of selective modulators of their activity. We recently described the structural requirements that led to the identification of potent and selective PRMT4 inhibitors spanning both the substrate and the cosubstrate pockets. The reanalysis of the data suggested a PRMT7 preferential binding for shorter derivatives and prompted us to extend these structural studies to PRMT9. Here, we report the identification of the first potent PRMT7/9 inhibitor and its binding mode to the two PRMT enzymes. Label-free quantification mass spectrometry confirmed significant inhibition of PRMT activity in cells. We also report the setup of an effective AlphaLISA assay to screen small molecule inhibitors of PRMT9.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


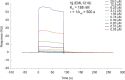


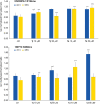
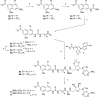
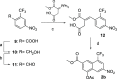
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information

