Base Dynamics in the Hha I Protein Binding Site
- PMID: 37561575
- PMCID: PMC10461302
- DOI: 10.1021/acs.jpcb.3c03687
Base Dynamics in the Hha I Protein Binding Site
Abstract
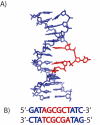
Protein-DNA interactions play an important role in numerous biological functions within the living cell. In many of these interactions, the DNA helix is significantly distorted upon protein-DNA complex formation. The HhaI restriction-modification system is one such system, where the methylation target is flipped out of the helix when bound to the methyltransferase. However, the base flipping mechanism is not well understood. The dynamics of the binding site of the HhaI methyltransferase and endonuclease (underlined) within the DNA oligomer [d(G1A2T3A4
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Backbone dynamics in the DNA HhaI protein binding site.J Am Chem Soc. 2008 Jul 16;130(28):9072-9. doi: 10.1021/ja801243d. Epub 2008 Jun 21. J Am Chem Soc. 2008. PMID: 18570423 Free PMC article.
-
Solid-state nuclear magnetic resonance spectroscopy studies of furanose ring dynamics in the DNA HhaI binding site.J Am Chem Soc. 2008 Jun 11;130(23):7305-14. doi: 10.1021/ja075775n. Epub 2008 May 20. J Am Chem Soc. 2008. PMID: 18489097
-
13C relaxation studies of the DNA target sequence for hhai methyltransferase reveal unique motional properties.Biochemistry. 2008 Jul 22;47(29):7617-25. doi: 10.1021/bi7020469. Epub 2008 Jun 26. Biochemistry. 2008. PMID: 18578505
-
Atomistic view of base flipping in DNA.Philos Trans A Math Phys Eng Sci. 2004 Jul 15;362(1820):1439-60. doi: 10.1098/rsta.2004.1383. Philos Trans A Math Phys Eng Sci. 2004. PMID: 15306460 Review.
-
DNA modification by methyltransferases.Curr Opin Struct Biol. 1995 Feb;5(1):4-10. doi: 10.1016/0959-440x(95)80003-j. Curr Opin Struct Biol. 1995. PMID: 7773746 Review.
Cited by
-
Twisting tetraplex DNA: A strand dynamics regulating i-motif function in diverse molecular crowding environments.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf500. doi: 10.1093/nar/gkaf500. Nucleic Acids Res. 2025. PMID: 40586303 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

