The All of Us Data and Research Center: Creating a Secure, Scalable, and Sustainable Ecosystem for Biomedical Research
- PMID: 37561600
- PMCID: PMC11157478
- DOI: 10.1146/annurev-biodatasci-122120-104825
The All of Us Data and Research Center: Creating a Secure, Scalable, and Sustainable Ecosystem for Biomedical Research
Abstract
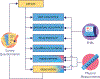
The All of Us Research Program's Data and Research Center (DRC) was established to help acquire, curate, and provide access to one of the world's largest and most diverse datasets for precision medicine research. Already, over 500,000 participants are enrolled in All of Us, 80% of whom are underrepresented in biomedical research, and data are being analyzed by a community of over 2,300 researchers. The DRC created this thriving data ecosystem by collaborating with engaged participants, innovative program partners, and empowered researchers. In this review, we first describe how the DRC is organized to meet the needs of this broad group of stakeholders. We then outline guiding principles, common challenges, and innovative approaches used to build the All of Us data ecosystem. Finally, we share lessons learned to help others navigate important decisions and trade-offs in building a modern biomedical data platform.
Keywords: data ecosystem; data integration; data privacy; diversity; electronic health records; precision medicine.
Figures



References
-
- The Cancer Genome Atlas Program - NCI. 2018. URL: https://www.cancer.gov/about-nci/organization/ccg/research/structural-ge... (Accessed 30 September 2022).
-
- ENCODE. n.d. URL: https://www.encodeproject.org/ (Accessed 5 October 2022).
-
- Human Genome Diversity Project. n.d. URL: https://hagsc.org/hgdp/ (Accessed 5 October 2022).
-
- CHARGE Consortium. n.d. URL: https://www.chargeconsortium.com/ (Accessed 5 October 2022).
Publication types
MeSH terms
Grants and funding
- OT2 OD026556/OD/NIH HHS/United States
- U2C OD023196/OD/NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- OT2 OD025277/OD/NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- OT2 OD026557/OD/NIH HHS/United States
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- OT2 OD035404/OD/NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- P30 DK092949/DK/NIDDK NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

