Zika-specific neutralizing antibodies targeting inter-dimer envelope epitopes
- PMID: 37561630
- PMCID: PMC10775418
- DOI: 10.1016/j.celrep.2023.112942
Zika-specific neutralizing antibodies targeting inter-dimer envelope epitopes
Abstract
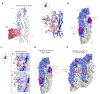
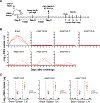
Zika virus (ZIKV) is an emerging pathogen that causes devastating congenital defects. The overlapping epidemiology and immunologic cross-reactivity between ZIKV and dengue virus (DENV) pose complex challenges to vaccine design, given the potential for antibody-dependent enhancement of disease. Therefore, classification of ZIKV-specific antibody targets is of notable value. From a ZIKV-infected rhesus macaque, we identify ZIKV-reactive B cells and isolate potent neutralizing monoclonal antibodies (mAbs) with no cross-reactivity to DENV. We group these mAbs into four distinct antigenic groups targeting ZIKV-specific cross-protomer epitopes on the envelope glycoprotein. Co-crystal structures of representative mAbs in complex with ZIKV envelope glycoprotein reveal envelope-dimer epitope and unique dimer-dimer epitope targeting. All four specificities are serologically identified in convalescent humans following ZIKV infection, and representative mAbs from all four groups protect against ZIKV replication in mice. These results provide key insights into ZIKV-specific antigenicity and have implications for ZIKV vaccine, diagnostic, and therapeutic development.
Keywords: CP: Immunology; X-ray crystallography; Zika virus; dengue virus; flavivirus; monoclonal antibodies; neutralizing antibodies; rhesus macaque; structural biology.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.J.K., V.D., G.D., K.M., N.M., D.H.B., M.G.J., R.S.S., and R.G.J. are named inventors on a PCT patent application WO 2019/209974 describing ZIKV neutralizing antibodies and their use. D.H.B. has received grants from Novavax and personal fees from IGM Biosciences. M.H., A.A., E.D., and B.J.D. are employees of Integral Molecular. B.J.D. is also a shareholder of the company.
Figures





References
-
- Cao-Lormeau VM, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. (2016). Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539. 10.1016/S0140-6736(16)00562-6. - DOI - PMC - PubMed
-
- Macnamara FN (1954). Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans. R. Soc. Trop. Med. Hyg. 48, 139–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

