Integrative splicing-quantitative-trait-locus analysis reveals risk loci for non-small-cell lung cancer
- PMID: 37562399
- PMCID: PMC10502736
- DOI: 10.1016/j.ajhg.2023.07.008
Integrative splicing-quantitative-trait-locus analysis reveals risk loci for non-small-cell lung cancer
Abstract
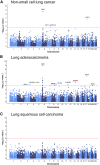
Splicing quantitative trait loci (sQTLs) have been demonstrated to contribute to disease etiology by affecting alternative splicing. However, the role of sQTLs in the development of non-small-cell lung cancer (NSCLC) remains unknown. Thus, we performed a genome-wide sQTL study to identify genetic variants that affect alternative splicing in lung tissues from 116 individuals of Chinese ancestry, which resulted in the identification of 1,385 sQTL-harboring genes (sGenes) containing 378,210 significant variant-intron pairs. A comprehensive characterization of these sQTLs showed that they were enriched in actively transcribed regions, genetic regulatory elements, and splicing-factor-binding sites. Moreover, sQTLs were largely distinct from expression quantitative trait loci (eQTLs) and showed significant enrichment in potential risk loci of NSCLC. We also integrated sQTLs into NSCLC GWAS datasets (13,327 affected individuals and 13,328 control individuals) by using splice-transcriptome-wide association study (spTWAS) and identified alternative splicing events in 19 genes that were significantly associated with NSCLC risk. By using functional annotation and experiments, we confirmed an sQTL variant, rs35861926, that reduced the risk of lung adenocarcinoma (rs35861926-T, OR = 0.88, 95% confidence interval [CI]: 0.82-0.93, p = 1.87 × 10-5) by promoting FARP1 exon 20 skipping to downregulate the expression level of the long transcript FARP1-011. Transcript FARP1-011 promoted the migration and proliferation of lung adenocarcinoma cells. Overall, our study provided informative lung sQTL resources and insights into the molecular mechanisms linking sQTL variants to NSCLC risk.
Keywords: FARP1 exon 20 skipping; non-small-cell lung cancer; risk loci; splice-transcriptome-wide association study; splicing quantitative trait locus.
Copyright © 2023 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Dai J., Lv J., Zhu M., Wang Y., Qin N., Ma H., He Y.Q., Zhang R., Tan W., Fan J., et al. Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir. Med. 2019;7:881–891. doi: 10.1016/S2213-2600(19)30144-4. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

