SF3B1 mutation-mediated sensitization to H3B-8800 splicing inhibitor in chronic lymphocytic leukemia
- PMID: 37562845
- PMCID: PMC10415613
- DOI: 10.26508/lsa.202301955
SF3B1 mutation-mediated sensitization to H3B-8800 splicing inhibitor in chronic lymphocytic leukemia
Abstract
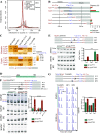
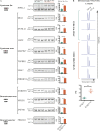
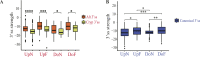
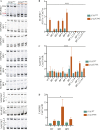
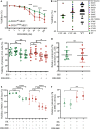
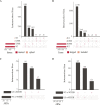
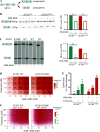
Splicing factor 3B subunit 1 (SF3B1) is involved in pre-mRNA branch site recognition and is the target of antitumor-splicing inhibitors. Mutations in SF3B1 are observed in 15% of patients with chronic lymphocytic leukemia (CLL) and are associated with poor prognosis, but their pathogenic mechanisms remain poorly understood. Using deep RNA-sequencing data from 298 CLL tumor samples and isogenic SF3B1 WT and K700E-mutated CLL cell lines, we characterize targets and pre-mRNA sequence features associated with the selection of cryptic 3' splice sites upon SF3B1 mutation, including an event in the MAP3K7 gene relevant for activation of NF-κB signaling. Using the H3B-8800 splicing modulator, we show, for the first time in CLL, cytotoxic effects in vitro in primary CLL samples and in SF3B1-mutated isogenic CLL cell lines, accompanied by major splicing changes and delayed leukemic infiltration in a CLL xenotransplant mouse model. H3B-8800 displayed preferential lethality towards SF3B1-mutated cells and synergism with the BCL2 inhibitor venetoclax, supporting the potential use of SF3B1 inhibitors as a novel therapeutic strategy in CLL.
© 2023 López-Oreja et al.
Conflict of interest statement
D Colomer reports grants and personal fees from H3 Biomedicine, Novartis, Incyte, AstraZeneca, and AbbVie.
Figures






References
-
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. (2016) Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 7: 10615. 10.1038/ncomms10615 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
