TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
- PMID: 37563110
- PMCID: PMC10415333
- DOI: 10.1038/s41392-023-01537-x
TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
Abstract
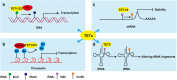
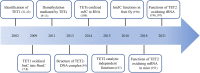
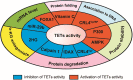
Ten-eleven translocation (TET) family proteins (TETs), specifically, TET1, TET2 and TET3, can modify DNA by oxidizing 5-methylcytosine (5mC) iteratively to yield 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC), and then two of these intermediates (5fC and 5caC) can be excised and return to unmethylated cytosines by thymine-DNA glycosylase (TDG)-mediated base excision repair. Because DNA methylation and demethylation play an important role in numerous biological processes, including zygote formation, embryogenesis, spatial learning and immune homeostasis, the regulation of TETs functions is complicated, and dysregulation of their functions is implicated in many diseases such as myeloid malignancies. In addition, recent studies have demonstrated that TET2 is able to catalyze the hydroxymethylation of RNA to perform post-transcriptional regulation. Notably, catalytic-independent functions of TETs in certain biological contexts have been identified, further highlighting their multifunctional roles. Interestingly, by reactivating the expression of selected target genes, accumulated evidences support the potential therapeutic use of TETs-based DNA methylation editing tools in disorders associated with epigenetic silencing. In this review, we summarize recent key findings in TETs functions, activity regulators at various levels, technological advances in the detection of 5hmC, the main TETs oxidative product, and TETs emerging applications in epigenetic editing. Furthermore, we discuss existing challenges and future directions in this field.
© 2023. West China Hospital, Sichuan University.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

