Applied machine learning as a driver for polymeric biomaterials design
- PMID: 37563117
- PMCID: PMC10415291
- DOI: 10.1038/s41467-023-40459-8
Applied machine learning as a driver for polymeric biomaterials design
Abstract
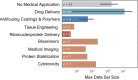
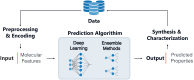
Polymers are ubiquitous to almost every aspect of modern society and their use in medical products is similarly pervasive. Despite this, the diversity in commercial polymers used in medicine is stunningly low. Considerable time and resources have been extended over the years towards the development of new polymeric biomaterials which address unmet needs left by the current generation of medical-grade polymers. Machine learning (ML) presents an unprecedented opportunity in this field to bypass the need for trial-and-error synthesis, thus reducing the time and resources invested into new discoveries critical for advancing medical treatments. Current efforts pioneering applied ML in polymer design have employed combinatorial and high throughput experimental design to address data availability concerns. However, the lack of available and standardized characterization of parameters relevant to medicine, including degradation time and biocompatibility, represents a nearly insurmountable obstacle to ML-aided design of biomaterials. Herein, we identify a gap at the intersection of applied ML and biomedical polymer design, highlight current works at this junction more broadly and provide an outlook on challenges and future directions.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Polykovskiy D, et al. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 2018;15:4398–4405. - PubMed
-
- Nguyen DH, Tsuda K. Generating reaction trees with cascaded variational autoencoders. J. Chem. Phys. 2022;156:044117. - PubMed
-
- Cai C, et al. Transfer learning for drug discovery. J. Med Chem. 2020;63:8683–8694. - PubMed
-
- Panteleev J, Gao H, Jia L. Recent applications of machine learning in medicinal chemistry. Bioorg. Med. Chem. Lett. 2018;28:2807–2815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

