Extravillous trophoblast cell lineage development is associated with active remodeling of the chromatin landscape
- PMID: 37563143
- PMCID: PMC10415281
- DOI: 10.1038/s41467-023-40424-5
Extravillous trophoblast cell lineage development is associated with active remodeling of the chromatin landscape
Abstract
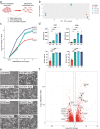
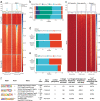
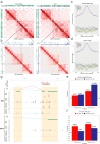
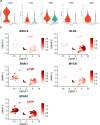
The extravillous trophoblast cell lineage is a key feature of placentation and successful pregnancy. Knowledge of transcriptional regulation driving extravillous trophoblast cell development is limited. Here, we map the transcriptome and epigenome landscape as well as chromatin interactions of human trophoblast stem cells and their transition into extravillous trophoblast cells. We show that integrating chromatin accessibility, long-range chromatin interactions, transcriptomic, and transcription factor binding motif enrichment enables identification of transcription factors and regulatory mechanisms critical for extravillous trophoblast cell development. We elucidate functional roles for TFAP2C, SNAI1, and EPAS1 in the regulation of extravillous trophoblast cell development. EPAS1 is identified as an upstream regulator of key extravillous trophoblast cell transcription factors, including ASCL2 and SNAI1 and together with its target genes, is linked to pregnancy loss and birth weight. Collectively, we reveal activation of a dynamic regulatory network and provide a framework for understanding extravillous trophoblast cell specification in trophoblast cell lineage development and human placentation.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The multifaceted roles of the transcriptional coactivator TAZ in extravillous trophoblast development of the human placenta.Proc Natl Acad Sci U S A. 2025 Apr 22;122(16):e2426385122. doi: 10.1073/pnas.2426385122. Epub 2025 Apr 14. Proc Natl Acad Sci U S A. 2025. PMID: 40228123 Free PMC article.
-
The transcriptional regulatory network modulating human trophoblast stem cells to extravillous trophoblast differentiation.Nat Commun. 2024 Feb 12;15(1):1285. doi: 10.1038/s41467-024-45669-2. Nat Commun. 2024. PMID: 38346993 Free PMC article.
-
ASCL2 reciprocally controls key trophoblast lineage decisions during hemochorial placenta development.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2016517118. doi: 10.1073/pnas.2016517118. Proc Natl Acad Sci U S A. 2021. PMID: 33649217 Free PMC article.
-
Early differentiation and gene expression characteristics of trophoblast lineages†.Biol Reprod. 2023 May 10;108(5):709-719. doi: 10.1093/biolre/ioad027. Biol Reprod. 2023. PMID: 36892411 Review.
-
Transcription factor networks in trophoblast development.Cell Mol Life Sci. 2022 Jun 3;79(6):337. doi: 10.1007/s00018-022-04363-6. Cell Mol Life Sci. 2022. PMID: 35657505 Free PMC article. Review.
Cited by
-
Super-Enhancers in Placental Development and Diseases.J Dev Biol. 2025 Apr 9;13(2):11. doi: 10.3390/jdb13020011. J Dev Biol. 2025. PMID: 40265369 Free PMC article. Review.
-
Aryl Hydrocarbon Receptor Activation Drives 2-Methoxy Estradiol Secretion in Human Trophoblast Stem Cell Development.bioRxiv [Preprint]. 2025 Jan 31:2024.08.27.609205. doi: 10.1101/2024.08.27.609205. bioRxiv. 2025. PMID: 39253430 Free PMC article. Preprint.
-
The multifaceted roles of the transcriptional coactivator TAZ in extravillous trophoblast development of the human placenta.Proc Natl Acad Sci U S A. 2025 Apr 22;122(16):e2426385122. doi: 10.1073/pnas.2426385122. Epub 2025 Apr 14. Proc Natl Acad Sci U S A. 2025. PMID: 40228123 Free PMC article.
-
Cited2 is a key regulator of placental development and plasticity.Bioessays. 2024 Aug;46(8):e2300118. doi: 10.1002/bies.202300118. Epub 2024 Jun 24. Bioessays. 2024. PMID: 38922923 Free PMC article. Review.
-
In vitro differentiation of macaque extravillous trophoblasts in a low oxygen environment.Placenta. 2025 Apr;163:16-28. doi: 10.1016/j.placenta.2025.02.014. Epub 2025 Feb 25. Placenta. 2025. PMID: 40024139
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

