Pharmacology of Compounds Targeting Cation-Chloride Cotransporter Physiology
- PMID: 37563251
- PMCID: PMC10823342
- DOI: 10.1007/164_2023_692
Pharmacology of Compounds Targeting Cation-Chloride Cotransporter Physiology
Abstract
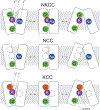
Transporters of the solute carrier family 12 (SLC12) carry inorganic cations such as Na+ and/or K+ alongside Cl across the plasma membrane of cells. These tightly coupled, electroneutral, transporters are expressed in almost all tissues/organs in the body where they fulfil many critical functions. The family includes two key transporters participating in salt reabsorption in the kidney: the Na-K-2Cl cotransporter-2 (NKCC2), expressed in the loop of Henle, and the Na-Cl cotransporter (NCC), expressed in the distal convoluted tubule. NCC and NKCC2 are the targets of thiazides and "loop" diuretics, respectively, drugs that are widely used in clinical medicine to treat hypertension and edema. Bumetanide, in addition to its effect as a loop diuretic, has recently received increasing attention as a possible therapeutic agent for neurodevelopmental disorders. This chapter also describes how over the past two decades, the pharmacology of Na+ independent transporters has expanded significantly to provide novel tools for research. This work has indeed led to the identification of compounds that are 100-fold to 1000-fold more potent than furosemide, the first described inhibitor of K-Cl cotransport, and identified compounds that possibly directly stimulate the function of the K-Cl cotransporter. Finally, the recent cryo-electron microscopy revolution has begun providing answers as to where and how pharmacological agents bind to and affect the function of the transporters.
Keywords: Bumetanide; Cation–chloride cotransporters; Chloride dye; Cryo-EM structure; High-throughput screening; Ion binding; Kidney; N-ethylmaleimide; Neurodevelopmental disorders; Protein–ligand complex; Thallium dyes; Thiazide and loop diuretics.
© 2023. The Author(s), under exclusive license to Springer Nature Switzerland AG.
Figures

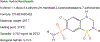


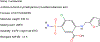


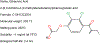

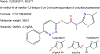
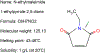


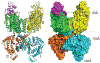





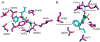

Similar articles
-
From Fish Physiology to Human Disease: The Discovery of the NCC, NKCC2, and the Cation-Coupled Chloride Cotransporters.Kidney360. 2024 Jan 1;5(1):133-141. doi: 10.34067/KID.0000000000000307. Epub 2023 Nov 16. Kidney360. 2024. PMID: 37968800 Free PMC article.
-
Chloride-Dependent Cation Transport via SLC12 Carriers at Atomic Resolution.Annu Rev Physiol. 2025 Feb;87(1):397-419. doi: 10.1146/annurev-physiol-022624-020130. Annu Rev Physiol. 2025. PMID: 39928503 Review.
-
Molecular physiology of cation-coupled Cl- cotransport: the SLC12 family.Pflugers Arch. 2004 Feb;447(5):580-93. doi: 10.1007/s00424-003-1066-3. Epub 2003 May 9. Pflugers Arch. 2004. PMID: 12739168 Review.
-
Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters.Physiol Rev. 2005 Apr;85(2):423-93. doi: 10.1152/physrev.00011.2004. Physiol Rev. 2005. PMID: 15788703 Review.
-
Abnormal reabsorption of Na+/CI- by the thiazide-inhibitable transporter of the distal convoluted tubule in Gitelman's syndrome.Am J Nephrol. 1997;17(2):103-11. doi: 10.1159/000169082. Am J Nephrol. 1997. PMID: 9096439
Cited by
-
Pharmacoepidemiology evaluation of bumetanide as a potential candidate for drug repurposing for Alzheimer's disease.Alzheimers Dement. 2024 Aug;20(8):5236-5246. doi: 10.1002/alz.13872. Epub 2024 Jun 21. Alzheimers Dement. 2024. PMID: 39030734 Free PMC article.
References
-
- Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL Jr., Jones DW, Materson BJ, Oparil S, Wright JT Jr., and Roccella EJ(2003) Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 42: 1206–1252 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

