Identification of human exTreg cells as CD16+CD56+ cytotoxic CD4+ T cells
- PMID: 37563308
- PMCID: PMC11022744
- DOI: 10.1038/s41590-023-01589-9
Identification of human exTreg cells as CD16+CD56+ cytotoxic CD4+ T cells
Abstract
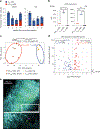
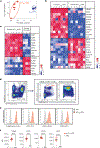
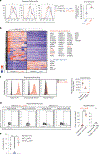
In atherosclerosis, some regulatory T (Treg) cells become exTreg cells. We crossed inducible Treg and exTreg cell lineage-tracker mice (FoxP3eGFP-Cre-ERT2ROSA26CAG-fl-stop-fl-tdTomato) to atherosclerosis-prone Apoe-/- mice, sorted Treg cells and exTreg cells and determined their transcriptomes by bulk RNA sequencing (RNA-seq). Genes that were differentially expressed between mouse Treg cells and exTreg cells and filtered for their presence in a human single-cell RNA-sequencing (scRNA-seq) panel identified exTreg cell signature genes as CST7, NKG7, GZMA, PRF1, TBX21 and CCL4. Projecting these genes onto the human scRNA-seq with CITE-seq data identified human exTreg cells as CD3+CD4+CD16+CD56+, which was validated by flow cytometry. Bulk RNA-seq of sorted human exTreg cells identified them as inflammatory and cytotoxic CD4+T cells that were significantly distinct from both natural killer and Treg cells. DNA sequencing for T cell receptor-β showed clonal expansion of Treg cell CDR3 sequences in exTreg cells. Cytotoxicity was functionally demonstrated in cell killing and CD107a degranulation assays, which identifies human exTreg cells as cytotoxic CD4+T cells.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

