A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology
- PMID: 37563457
- PMCID: PMC10505560
- DOI: 10.1038/s41477-023-01489-8
A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology
Erratum in
-
Author Correction: A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology.Nat Plants. 2024 Jan;10(1):193. doi: 10.1038/s41477-024-01618-x. Nat Plants. 2024. PMID: 38182752 Free PMC article. No abstract available.
Abstract
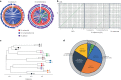
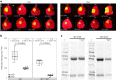
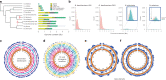
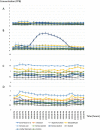
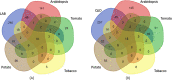
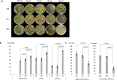
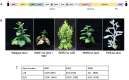
Nicotiana benthamiana is an invaluable model plant and biotechnology platform with a ~3 Gb allotetraploid genome. To further improve its usefulness and versatility, we have produced high-quality chromosome-level genome assemblies, coupled with transcriptome, epigenome, microRNA and transposable element datasets, for the ubiquitously used LAB strain and a related wild accession, QLD. In addition, single nucleotide polymorphism maps have been produced for a further two laboratory strains and four wild accessions. Despite the loss of five chromosomes from the ancestral tetraploid, expansion of intergenic regions, widespread segmental allopolyploidy, advanced diploidization and evidence of recent bursts of Copia pseudovirus (Copia) mobility not seen in other Nicotiana genomes, the two subgenomes of N. benthamiana show large regions of synteny across the Solanaceae. LAB and QLD have many genetic, metabolic and phenotypic differences, including disparate RNA interference responses, but are highly interfertile and amenable to genome editing and both transient and stable transformation. The LAB/QLD combination has the potential to be as useful as the Columbia-0/Landsberg errecta partnership, utilized from the early pioneering days of Arabidopsis genomics to today.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Narayan RKJ. Nuclear DNA changes, genome differentiation and evolution in Nicotiana (Solanaceae) Plant Syst. Evol. 1987;157:161–180. doi: 10.1007/BF00936195. - DOI
-
- Marks CE, Ladiges PY, Newbigin E. Karyotypic variation in Nicotiana section Suaveolentes. Genet. Resour. Crop Evol. 2011;58:797–803. doi: 10.1007/s10722-011-9724-3. - DOI
-
- Bally J, et al. Nicotiana paulineana, a new Australian species in Nicotiana section Suaveolentes. Aust. Syst. Bot. 2021;34:477–484. doi: 10.1071/SB20025. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

