The 18S rRNA genes of Haemoproteus (Haemosporida, Apicomplexa) parasites from European songbirds with remarks on improved parasite diagnostics
- PMID: 37563610
- PMCID: PMC10416517
- DOI: 10.1186/s12936-023-04661-9
The 18S rRNA genes of Haemoproteus (Haemosporida, Apicomplexa) parasites from European songbirds with remarks on improved parasite diagnostics
Abstract
Background: The nuclear ribosomal RNA genes of Plasmodium parasites are assumed to evolve according to a birth-and-death model with new variants originating by duplication and others becoming deleted. For some Plasmodium species, it has been shown that distinct variants of the 18S rRNA genes are expressed differentially in vertebrate hosts and mosquito vectors. The central aim was to evaluate whether avian haemosporidian parasites of the genus Haemoproteus also have substantially distinct 18S variants, focusing on lineages belonging to the Haemoproteus majoris and Haemoproteus belopolskyi species groups.
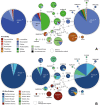
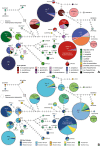
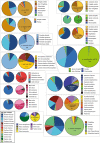
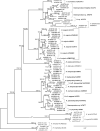
Methods: The almost complete 18S rRNA genes of 19 Haemoproteus lineages of the subgenus Parahaemoproteus, which are common in passeriform birds from the Palaearctic, were sequenced. The PCR products of 20 blood and tissue samples containing 19 parasite lineages were subjected to molecular cloning, and ten clones in mean were sequenced each. The sequence features were analysed and phylogenetic trees were calculated, including sequence data published previously from eight additional Parahaemoproteus lineages. The geographic and host distribution of all 27 lineages was visualised as CytB haplotype networks and pie charts. Based on the 18S sequence data, species-specific oligonucleotide probes were designed to target the parasites in host tissue by in situ hybridization assays.
Results: Most Haemoproteus lineages had two or more variants of the 18S gene like many Plasmodium species, but the maximum distances between variants were generally lower. Moreover, unlike in most mammalian and avian Plasmodium species, the 18S sequences of all but one parasite lineage clustered into reciprocally monophyletic clades. Considerably distinct 18S clusters were only found in Haemoproteus tartakovskyi hSISKIN1 and Haemoproteus sp. hROFI1. The presence of chimeric 18S variants in some Haemoproteus lineages indicates that their ribosomal units rather evolve in a semi-concerted fashion than according to a strict model of birth-and-death evolution.
Conclusions: Parasites of the subgenus Parahaemoproteus contain distinct 18S variants, but the intraspecific variability is lower than in most mammalian and avian Plasmodium species. The new 18S data provides a basis for more thorough investigations on the development of Haemoproteus parasites in host tissue using in situ hybridization techniques targeting specific parasite lineages.
Keywords: Birth-and-death evolution; Parahaemoproteus; Ribosomal genes; Semi-concerted evolution.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Haemoproteus tartakovskyi and Plasmodium relictum (Haemosporida, Apicomplexa) differentially express distinct 18S rRNA gene variants in bird hosts and dipteran vectors.Parasit Vectors. 2025 Feb 20;18(1):63. doi: 10.1186/s13071-025-06696-0. Parasit Vectors. 2025. PMID: 39979994 Free PMC article.
-
The nuclear 18S ribosomal DNAs of avian haemosporidian parasites.Malar J. 2019 Sep 3;18(1):305. doi: 10.1186/s12936-019-2940-6. Malar J. 2019. PMID: 31481072 Free PMC article.
-
Avian haemosporidian parasites of accipitriform raptors.Malar J. 2022 Jan 5;21(1):14. doi: 10.1186/s12936-021-04019-z. Malar J. 2022. PMID: 34986864 Free PMC article.
-
Keys to the avian Haemoproteus parasites (Haemosporida, Haemoproteidae).Malar J. 2022 Sep 19;21(1):269. doi: 10.1186/s12936-022-04235-1. Malar J. 2022. PMID: 36123731 Free PMC article. Review.
-
The influence of the host sex on parasitemia of parasite lineages belonging to Haemoproteus majoris in a natural bird community.Parasitol Res. 2023 Apr;122(4):895-901. doi: 10.1007/s00436-023-07793-8. Epub 2023 Feb 13. Parasitol Res. 2023. PMID: 36781472 Review.
Cited by
-
Imprudent use of MalAvi names biases the estimation of parasite diversity of avian haemosporidians.PLoS Pathog. 2025 Feb 5;21(2):e1012911. doi: 10.1371/journal.ppat.1012911. eCollection 2025 Feb. PLoS Pathog. 2025. PMID: 39908324 Free PMC article. No abstract available.
-
Haemoproteus tartakovskyi and Plasmodium relictum (Haemosporida, Apicomplexa) differentially express distinct 18S rRNA gene variants in bird hosts and dipteran vectors.Parasit Vectors. 2025 Feb 20;18(1):63. doi: 10.1186/s13071-025-06696-0. Parasit Vectors. 2025. PMID: 39979994 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

