CIS deletion by CRISPR/Cas9 enhances human primary natural killer cell functions against allogeneic glioblastoma
- PMID: 37563692
- PMCID: PMC10413513
- DOI: 10.1186/s13046-023-02770-6
CIS deletion by CRISPR/Cas9 enhances human primary natural killer cell functions against allogeneic glioblastoma
Abstract
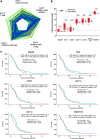
Background: Glioblastoma (GBM) is the most common malignant brain tumor and has "immunologically cold" features. Changing GBM to an "immunologically hot" tumor requires a strong trigger that induces initial immune responses in GBM. Allogeneic natural killer cells (NKCs) have gained considerable attention as promising immunotherapeutic tools against cancer, where gene-edited NKCs would result in effective anti-cancer treatment. The present study focused on the immune checkpoint molecule cytokine-inducible SH2-containing protein (CISH, or CIS) as a critical negative regulator in NKCs.
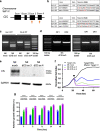
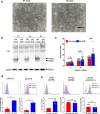
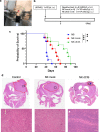
Methods: The GBM tumor environment featured with immunological aspect was analyzed with Cancer immunogram and GlioVis. We generated human primary CIS-deleted NKCs (NK dCIS) using clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) with single guide RNA targeting genome sites on CIS coding exons. The genome-edited NKCs underwent microarray with differential expression analysis and gene set enrichment analysis (GSEA). The anti-GBM activity of the genome-edited NKCs was evaluated by apoptosis induction effects against allogeneic GBM cells and spheroids. We further detected in vivo antitumor effects using xenograft brain tumor mice.
Results: We successfully induced human CIS-deleted NKCs (NK dCIS) by combining our specific human NKC expansion method available for clinical application and genome editing technology. CIS gene-specific guide RNA/Cas9 protein complex suppressed CIS expression in the expanded NKCs with high expansion efficacy. Comprehensive gene expression analysis demonstrated increased expression of 265 genes and decreased expression of 86 genes in the NK dCIS. Gene set enrichment analysis revealed that the enriched genes were involved in NKC effector functions. Functional analysis revealed that the NK dCIS had increased interferon (IFN)ɤ and tumor necrosis factor (TNF) production. CIS deletion enhanced NKC-mediated apoptosis induction against allogeneic GBM cells and spheroids. Intracranial administration of the allogeneic NKCs prolonged the overall survival of xenograft brain tumor mice. Furthermore, the NK dCIS extended the overall survival of the mice.
Conclusion: The findings demonstrated the successful induction of human primary NK dCIS with CRISPR/Cas9 with efficient expansion. CIS deletion enhanced the NKC-mediated anti-tumor effects in allogeneic GBM and could be a promising immunotherapeutic alternative for patients with GBM.
Keywords: CIS; CRISPR/Cas9; Glioblastoma; Natural killer cell; Peripheral blood.
© 2023. Italian National Cancer Institute ‘Regina Elena’.
Conflict of interest statement
T.N. and M.N. are employees of Grandsoul Research Institute for Immunology, Inc. and Clinic Grandsoul Nara, respectively. The other authors declare no conflicts of interest. There are no competing financial interests in relation to the work described.
Figures




References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

