Purifying selection against spurious splicing signals contributes to the base composition evolution of the polypyrimidine tract
- PMID: 37564008
- PMCID: PMC10946897
- DOI: 10.1111/jeb.14205
Purifying selection against spurious splicing signals contributes to the base composition evolution of the polypyrimidine tract
Abstract
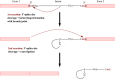
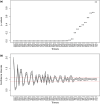
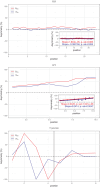
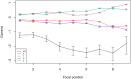
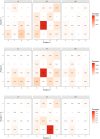
Among eukaryotes, the major spliceosomal pathway is highly conserved. While long introns may contain additional regulatory sequences, the ones in short introns seem to be nearly exclusively related to splicing. Although these regulatory sequences involved in splicing are well-characterized, little is known about their evolution. At the 3' end of introns, the splice signal nearly universally contains the dimer AG, which consists of purines, and the polypyrimidine tract upstream of this 3' splice signal is characterized by over-representation of pyrimidines. If the over-representation of pyrimidines in the polypyrimidine tract is also due to avoidance of a premature splicing signal, we hypothesize that AG should be the most under-represented dimer. Through the use of DNA-strand asymmetry patterns, we confirm this prediction in fruit flies of the genus Drosophila and by comparing the asymmetry patterns to a presumably neutrally evolving region, we quantify the selection strength acting on each motif. Moreover, our inference and simulation method revealed that the best explanation for the base composition evolution of the polypyrimidine tract is the joint action of purifying selection against a spurious 3' splice signal and the selection for pyrimidines. Patterns of asymmetry in other eukaryotes indicate that avoidance of premature splicing similarly affects the nucleotide composition in their polypyrimidine tracts.
Keywords: Drosophila; intron evolution; polypyrimidine tract; selective constraint; short intron; splicing motifs.
© 2023 The Authors. Journal of Evolutionary Biology published by John Wiley & Sons Ltd on behalf of European Society for Evolutionary Biology.
Conflict of interest statement
The authors declare no competing interests.
Figures

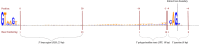



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

