Regulation of cellular contractile force, shape and migration of fibroblasts by oncogenes and Histone deacetylase 6
- PMID: 37564130
- PMCID: PMC10411354
- DOI: 10.3389/fmolb.2023.1197814
Regulation of cellular contractile force, shape and migration of fibroblasts by oncogenes and Histone deacetylase 6
Abstract
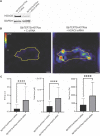
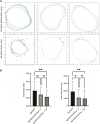
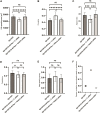
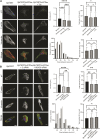
The capacity of cells to adhere to, exert forces upon and migrate through their surrounding environment governs tissue regeneration and cancer metastasis. The role of the physical contractile forces that cells exert in this process, and the underlying molecular mechanisms are not fully understood. We, therefore, aimed to clarify if the extracellular forces that cells exert on their environment and/or the intracellular forces that deform the cell nucleus, and the link between these forces, are defective in transformed and invasive fibroblasts, and to indicate the underlying molecular mechanism of control. Confocal, Epifluorescence and Traction force microscopy, followed by computational analysis, showed an increased maximum contractile force that cells apply on their environment and a decreased intracellular force on the cell nucleus in the invasive fibroblasts, as compared to normal control cells. Loss of HDAC6 activity by tubacin-treatment and siRNA-mediated HDAC6 knockdown also reversed the reduced size and more circular shape and defective migration of the transformed and invasive cells to normal. However, only tubacin-mediated, and not siRNA knockdown reversed the increased force of the invasive cells on their surrounding environment to normal, with no effects on nuclear forces. We observed that the forces on the environment and the nucleus were weakly positively correlated, with the exception of HDAC6 siRNA-treated cells, in which the correlation was weakly negative. The transformed and invasive fibroblasts showed an increased number and smaller cell-matrix adhesions than control, and neither tubacin-treatment, nor HDAC6 knockdown reversed this phenotype to normal, but instead increased it further. This highlights the possibility that the control of contractile force requires separate functions of HDAC6, than the control of cell adhesions, spreading and shape. These data are consistent with the possibility that defective force-transduction from the extracellular environment to the nucleus contributes to metastasis, via a mechanism that depends upon HDAC6. To our knowledge, our findings present the first correlation between the cellular forces that deforms the surrounding environment and the nucleus in fibroblasts, and it expands our understanding of how cells generate contractile forces that contribute to cell invasion and metastasis.
Keywords: Histone deacetylase 6; Traction force microscopy; cell adhesion; cellular contractile forces; fibroblasts; intracellular forces on nucleus; metastasis; oncogenes.
Copyright © 2023 López-Guajardo, Zafar, Al Hennawi, Rossi, Alrwaili, Medcalf, Dunning, Nordgren, Pettersson, Estabrook, Hawkins and Gad.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Metastasising Fibroblasts Show an HDAC6-Dependent Increase in Migration Speed and Loss of Directionality Linked to Major Changes in the Vimentin Interactome.Int J Mol Sci. 2022 Feb 10;23(4):1961. doi: 10.3390/ijms23041961. Int J Mol Sci. 2022. PMID: 35216078 Free PMC article.
-
The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells.Rep Prog Phys. 2019 Jun;82(6):064602. doi: 10.1088/1361-6633/ab1628. Epub 2019 Apr 4. Rep Prog Phys. 2019. PMID: 30947151 Review.
-
Rho GTPases link cellular contractile force to the density and distribution of nanoscale adhesions.FASEB J. 2012 Jun;26(6):2374-82. doi: 10.1096/fj.11-195800. Epub 2012 Feb 27. FASEB J. 2012. PMID: 22371528
-
Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents.Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):20003-8. doi: 10.1073/pnas.1013754107. Epub 2010 Oct 29. Proc Natl Acad Sci U S A. 2010. PMID: 21037108 Free PMC article.
-
Forces in cell locomotion.Biochem Soc Symp. 1999;65:299-314. Biochem Soc Symp. 1999. PMID: 10320946 Review.
References
LinkOut - more resources
Full Text Sources

