Lysine N-methyltransferase SETD7 promotes bladder cancer progression and immune escape via STAT3/PD-L1 cascade
- PMID: 37564199
- PMCID: PMC10411476
- DOI: 10.7150/ijbs.87182
Lysine N-methyltransferase SETD7 promotes bladder cancer progression and immune escape via STAT3/PD-L1 cascade
Abstract
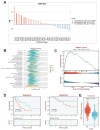
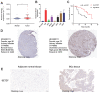
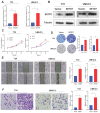
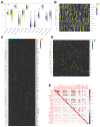
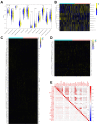
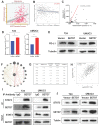
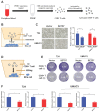
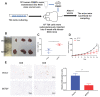
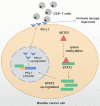
Background: The immunotherapy sensitivity of patients with bladder cancer (BCa) remains low. As the role of protein methylation in tumorigenesis and development becomes clearer, the role of lysine N-methyltransferase SET domain containing 7 (SETD7) in the progression and immune escape of BCa is worth studying. Methods: The correlation between lysine methyltransferase family and prognosis or immunotheray sensitivity of BCa patients were analyzed, and SETD7 was screened out because of the significant correlation between its expression and survival data or immunotherapy sensitivity. The expression of SETD7 in BCa tissues and cell lines were explored. The functions of SETD7 were investigated by proliferation and migration assays. The role of SETD7 in BCa immune escape was validated by analyzing the correlation between SETD7 expression and tumor microenvironment (TME)-related indicators. The results were further confirmed by conducting BCa cell-CD8+ T cell co-culture assays and tumorigenesis experiment in human immune reconstitution NOG mice (HuNOG mice). Bioinformatic prediction, CO-IP, qRT-PCR, and western blot were used to validate the SETD7/STAT3/PD-L1 cascade. Results: SETD7 was highly expressed in BCa, and it was positively associated with high histological grade and worse prognosis. SETD7 promoted the proliferation and migration of BCa cells. The results of bioinformatics, in vitro co-culture, and in vivo tumorigenesis assays showed that SETD7 could inhibit the chemotoxis and cytotoxicity of CD8+ T cells in BCa TME. Mechanistically, bioinformatics analysis, CO-IP assay, qRT-PCR, and western blot results indicated that SETD7 could increase the expression of PD-L1 via binding and promoting STAT3. Conclusions: Taken together, SETD7 indicated poor prognosis and promoted the progression and immune escape of BCa cells. It has great potential to act as a new indicator for BCa diagnosis and treatment, especially immunotherapy.
Keywords: SETD7; bladder cancer; immune escape; migration; proliferation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
HNRNPL induced circFAM13B increased bladder cancer immunotherapy sensitivity via inhibiting glycolysis through IGF2BP1/PKM2 pathway.J Exp Clin Cancer Res. 2023 Feb 6;42(1):41. doi: 10.1186/s13046-023-02614-3. J Exp Clin Cancer Res. 2023. PMID: 36747239 Free PMC article.
-
Cancer-associated Fibroblast-derived Extracellular Vesicles Mediate Immune Escape of Bladder Cancer via PD-L1/PD-1 Expression.Endocr Metab Immune Disord Drug Targets. 2023;23(11):1410-1420. doi: 10.2174/1871530323666230228124125. Endocr Metab Immune Disord Drug Targets. 2023. PMID: 36852791
-
Membrane palmitoylated protein MPP1 inhibits immune escape by regulating the USP12/ CCL5 axis in urothelial carcinoma.Int Immunopharmacol. 2025 Jan 27;146:113802. doi: 10.1016/j.intimp.2024.113802. Epub 2024 Dec 18. Int Immunopharmacol. 2025. PMID: 39700963
-
Lysine methyltransferase SETD7 in cancer: functions, molecular mechanisms and therapeutic implications.Mol Biol Rep. 2025 Apr 15;52(1):389. doi: 10.1007/s11033-025-10494-3. Mol Biol Rep. 2025. PMID: 40232640 Review.
-
The role of SET domain containing lysine methyltransferase 7 in tumorigenesis and development.Cell Cycle. 2023 Feb;22(3):269-275. doi: 10.1080/15384101.2022.2122257. Epub 2022 Sep 13. Cell Cycle. 2023. PMID: 36101480 Free PMC article. Review.
Cited by
-
Blockade of the STAT3/BCL-xL Axis Leads to the Cytotoxic and Cisplatin-Sensitizing Effects of Fucoxanthin, a Marine-Derived Carotenoid, on Human Bladder Urothelial Carcinoma Cells.Mar Drugs. 2025 Jan 22;23(2):54. doi: 10.3390/md23020054. Mar Drugs. 2025. PMID: 39997178 Free PMC article.
-
Programmed death receptor (PD-)1/PD-ligand (L)1 in urological cancers : the "all-around warrior" in immunotherapy.Mol Cancer. 2024 Sep 2;23(1):183. doi: 10.1186/s12943-024-02095-8. Mol Cancer. 2024. PMID: 39223527 Free PMC article. Review.
-
Lysine methylation modifications in tumor immunomodulation and immunotherapy: regulatory mechanisms and perspectives.Biomark Res. 2024 Jul 30;12(1):74. doi: 10.1186/s40364-024-00621-w. Biomark Res. 2024. PMID: 39080807 Free PMC article. Review.
-
Leveraging programmed cell death signature to predict clinical outcome and immunotherapy benefits in postoperative bladder cancer.Sci Rep. 2024 Oct 3;14(1):22976. doi: 10.1038/s41598-024-73571-w. Sci Rep. 2024. PMID: 39363008 Free PMC article.
-
ACAT2 suppresses the ubiquitination of YAP1 to enhance the proliferation and metastasis ability of gastric cancer via the upregulation of SETD7.Cell Death Dis. 2024 Apr 26;15(4):297. doi: 10.1038/s41419-024-06666-x. Cell Death Dis. 2024. PMID: 38670954 Free PMC article.
References
-
- Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol. 2017;71:96–108. - PubMed
-
- van Rhijn BW, Burger M, Lotan Y, Solsona E, Stief CG, Sylvester RJ. et al. Recurrence and progression of disease in non-muscle-invasive bladder cancer: from epidemiology to treatment strategy. Eur Urol. 2009;56:430–42. - PubMed
-
- Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder Cancer: A Review. Jama. 2020;324:1980–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

