Integrative multi-omics analyses unravel the immunological implication and prognostic significance of CXCL12 in breast cancer
- PMID: 37564657
- PMCID: PMC10410148
- DOI: 10.3389/fimmu.2023.1188351
Integrative multi-omics analyses unravel the immunological implication and prognostic significance of CXCL12 in breast cancer
Abstract
Background: CXCL12 is a vital factor in physiological and pathological processes, by inducing migration of multiple cells. We aimed to comprehensively detect the role of CXCL12 in breast cancer, and explore novel CXCL12-related biomarkers through integrative multi-omics analyses to build a powerful prognostic model for breast cancer patients.
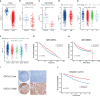
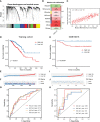
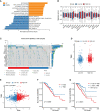
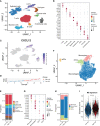
Methods: Immunohistochemistry analysis of the tissue microarray was performed to evaluate the correlation between CXCL12 expression levels and breast cancer patient outcomes. Combined single-nucleus and spatial transcriptomics data was used to uncover the expression distribution of CXCL12 in breast cancer microenvironment. CXCL12-related genes were identified by WGCNA analysis. Univariate Cox and LASSO regression analyses were then conducted to screen prognostic genes from above CXCL12-related genes, followed by the construction of the CXCL12-related prognostic signature, identification of risk groups, and external validation of the prognostic signature. Analyses of biological function, mutation landscape, immune checkpoint genes and immune cells, were performed to further reveal the differences between high/low-risk groups. Paired single-cell RNA-seq and bulk RNA-seq were analyzed to further disclose the association between the risk score and the complex tumor immune microenvironment. To screen potential therapeutic agents for breast cancer patients, analyses of gene-drug correlation and sensitivity to immunotherapy were conducted.
Results: High expression of CXCL12 was linked with a prolonged survival in breast cancer. A total of 402 genes were identified by WGCNA analysis and 11 genes, covering VAT1L, TMEM92, SDC1, RORB, PCSK9, NRN1, NACAD, JPH3, GJA1, BMP8B and ADAMTS2, were screened as the candidate prognostic genes. Next, the prognostic signature was built and validated using these genes to predict the outcomes of breast cancers. The high-risk group patients exhibited significantly inferior prognoses. The combination of the risk score and tumor mutational burden (TMB) had remarkably improved performance in predicting patient outcomes. Besides, high-risk group patients showed higher infiltration of M2-like macrophages. Finally, several potential anticancer drugs were identified. The high-risk group patients were more sensitive to immunotherapy but resistant to docetaxel.
Conclusions: CXCL12 has important immunological implication and prognostic significance in breast cancer. The CXCL12-related prognostic model could well predict the prognosis and treatment response of breast cancers.
Keywords: CXCL12; breast cancer; drug screening; immune landscape; prognostic signature; single-cell RNA-seq.
Copyright © 2023 Gao, Fang, Yuan, Sun and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

