Compartment-driven imprinting of intestinal CD4 T cells in inflammatory bowel disease and homeostasis
- PMID: 37565620
- PMCID: PMC10719222
- DOI: 10.1093/cei/uxad095
Compartment-driven imprinting of intestinal CD4 T cells in inflammatory bowel disease and homeostasis
Abstract
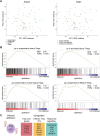
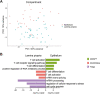
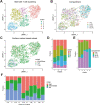
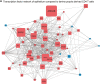
The mucosal immune system is implicated in the etiology and progression of inflammatory bowel diseases. The lamina propria and epithelium of the gut mucosa constitute two separate compartments, containing distinct T-cell populations. Human CD4 T-cell programming and regulation of lamina propria and epithelium CD4 T cells, especially during inflammation, remain incompletely understood. We performed flow cytometry, bulk, and single-cell RNA-sequencing to profile ileal lamina propria and intraepithelial CD4 T cells (CD4CD8αα, regulatory T cells (Tregs), CD69- and CD69high Trm T cells) in controls and Crohn's disease (CD) patients (paired non-inflamed and inflamed). Inflammation results in alterations of the CD4 T-cell population with a pronounced increase in Tregs and migrating/infiltrating cells. On a transcriptional level, inflammation within the epithelium induced T-cell activation, increased IFNγ responses, and an effector Treg profile. Conversely, few transcriptional changes within the lamina propria were observed. Key regulators including the chromatin remodelers ARID4B and SATB1 were found to drive compartment-specific transcriptional programming of CD4 T(reg) cells. In summary, inflammation in CD patients primarily induces changes within the epithelium and not the lamina propria. Additionally, there is compartment-specific CD4 T-cell imprinting, driven by shared regulators, between the lamina propria and the epithelium. The main consequence of intraepithelial adaptation, irrespective of inflammation, seems to be an overall dampening of broad (pro-inflammatory) responses and tight regulation of lifespan. These data suggest differential regulation of the lamina propria and epithelium, with a specific regulatory role in the inflamed epithelium.
Keywords: CD4 T cells; RNA-sequencing; adaptation; inflammatory bowel diseases; small intestine.
© The Author(s) 2023. Published by Oxford University Press on behalf of the British Society for Immunology.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

