Unrecognized risk of perfluorooctane sulfonate in promoting conjugative transfers of bacterial antibiotic resistance genes
- PMID: 37565764
- PMCID: PMC10537727
- DOI: 10.1128/aem.00533-23
Unrecognized risk of perfluorooctane sulfonate in promoting conjugative transfers of bacterial antibiotic resistance genes
Abstract
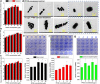
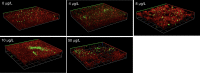
Antibiotic resistance is a major global health crisis facing humanity, with horizontal gene transfer (HGT) as a principal dissemination mechanism in the natural and clinical environments. Perfluoroalkyl substances (PFASs) are emerging contaminants of global concern due to their high persistence in the environment and adverse effects on humans. However, it is unknown whether PFASs affect the HGT of bacterial antibiotic resistance. Using a genetically engineered Escherichia coli MG1655 as the donor of plasmid-encoded antibiotic resistance genes (ARGs), E. coli J53 and soil bacterial community as two different recipients, this study demonstrated that the conjugation frequency of ARGs between two E. coli strains was (1.45 ± 0.17) × 10-5 and perfluorooctane sulfonate (PFOS) at environmentally relevant concentrations (2-50 μg L-1) increased conjugation transfer between E. coli strains by up to 3.25-fold. Increases in reactive oxygen species production, cell membrane permeability, biofilm formation capacity, and cell contact in two E. coli strains were proposed as major promotion mechanisms from PFOS exposure. Weighted gene co-expression network analysis of transcriptome data identified a series of candidate genes whose expression changes could contribute to the increase in conjugation transfer induced by PFOS. Furthermore, PFOS also generally increased the ARG transfer into the studied soil bacterial community, although the uptake ability of different community members of the plasmid either increased or decreased upon PFOS exposure depending on specific bacterial taxa. Overall, this study reveals an unrecognized risk of PFOS in accelerating the dissemination of antibiotic resistance. IMPORTANCE Perfluoroalkyl substances (PFASs) are emerging contaminants of global concern due to their high persistence in the environment and adverse health effects. Although the influence of environmental pollutants on the spread of antibiotic resistance, one of the biggest threats to global health, has attracted increasing attention in recent years, it is unknown whether environmental residues of PFASs affect the dissemination of bacterial antibiotic resistance. Considering PFASs, often called "forever" compounds, have significantly higher environmental persistence than most emerging organic contaminants, exploring the effect of PFASs on the spread of antibiotic resistance is more environmentally relevant and has essential ecological and health significance. By systematically examining the influence of perfluorooctane sulfonate on the antibiotic resistance gene conjugative transfer, not only at the single-strain level but also at the community level, this study has uncovered an unrecognized risk of PFASs in promoting conjugative transfers of bacterial antibiotic resistance genes, which could be incorporated into the risk assessment framework of PFASs.
Keywords: PFASs; antibiotic resistance; bacterial community; emerging pollutant; horizontal gene transfer; risk assessment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Achalapong S, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Babin F-X, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Berkley JA, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Clotaire Donatien R, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Duong Bich T, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garcia C, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Hsia Y, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Jenney AWJ, Khorana M, Khusuwan S, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Lim K, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Manoharan A, Marks F, May J, Mayxay M, Mturi N, Munera-Huertas T, Musicha P, Musila LA, Mussi-Pinhata MM, Naidu RN, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Ochoa TJ, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Ounchanum P, Pak GD, Paredes JL, Peleg AY, Perrone C, Phe T, Phommasone K, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Rattanavong S, Riddell A, Roberts T, Robotham JV, Roca A, Rosenthal VD, Rudd KE, Russell N, Sader HS, Saengchan W, Schnall J, Scott JAG, Seekaew S, Sharland M, Shivamallappa M, Sifuentes-Osornio J, Simpson AJ, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Tigoi C, Turner C, Turner P, van Doorn HR, Velaphi S, Vongpradith A, Vongsouvath M, Vu H, Walsh T, Walson JL, Waner S, Wangrangsimakul T, Wannapinij P, Wozniak T, Young Sharma TEMW, Yu KC, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 - DOI - PMC - PubMed
-
- Sun M, Arevalo E, Strynar M, Lindstrom A, Richardson M, Kearns B, Pickett A, Smith C, Knappe DRU. 2016. Legacy and emerging perfluoroalkyl substances are important drinking water contaminants in the cape fear river watershed of North Carolina. Environ Sci Technol Lett 3:415–419. doi: 10.1021/acs.estlett.6b00398 - DOI
-
- USEPA . 2016. Drinking water health Advisories for PFOA and PFOS - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

