Easy, Robust, and Repeatable Online Acid Cleavage of Proteins in Mobile Phase for Fast Quantitative LC-MS Bottom-Up Protein Analysis─Application for Ricin Detection
- PMID: 37565982
- PMCID: PMC10448442
- DOI: 10.1021/acs.analchem.3c01772
Easy, Robust, and Repeatable Online Acid Cleavage of Proteins in Mobile Phase for Fast Quantitative LC-MS Bottom-Up Protein Analysis─Application for Ricin Detection
Abstract
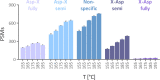
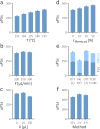
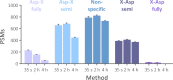
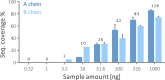
Sample preparation involving the cleavage of proteins into peptides is the first critical step for successful bottom-up proteomics and protein analyses. Time- and labor-intensiveness are among the bottlenecks of the commonly used methods for protein sample preparation. Here, we report a fast online method for postinjection acid cleavage of proteins directly in the mobile phase typically used for LC-MS analyses in proteomics. The chemical cleavage is achieved in 0.1% formic acid within 35 s in a capillary heated to 195 °C installed upstream of the analytical column, enabling the generated peptides to be separated. The peptides generated by the optimized method covered the entire sequence except for one amino acid of trastuzumab used for the method development. The qualitative results are extraordinarily stable, even over a long period of time. Moreover, the method is also suitable for accurate and repeatable quantification. The procedure requires only one manual step, significantly decreasing sample transfer losses. To demonstrate its practical utility, we tested the method for the fast detection of ricin. Ricin can be unambiguously identified from an injection of 10 ng, and the results can be obtained within 7-8 min after receiving a suspicious sample. Because no sophisticated accessories and no additional reagents are needed, the method can be seamlessly transferred to any laboratory for high-throughput proteomic workflows.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Capelo J. L.; Carreira R.; Diniz M.; Fernandes L.; Galesio M.; Lodeiro C.; Santos H. M.; Vale G. Overview on modern approaches to speed up protein identification workflows relying on enzymatic cleavage and mass spectrometry-based techniques. Anal. Chim. Acta 2009, 650, 151–159. 10.1016/j.aca.2009.07.034. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

