The adenylate cyclase toxin RTX domain follows a series templated folding mechanism with implications for toxin activity
- PMID: 37567473
- PMCID: PMC10511787
- DOI: 10.1016/j.jbc.2023.105150
The adenylate cyclase toxin RTX domain follows a series templated folding mechanism with implications for toxin activity
Abstract
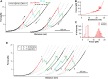
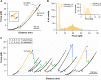
Folding of the Repeats-in-toxin (RTX) domain of the bacterial adenylate cyclase toxin-hemolysin (CyaA) is critical to its toxin activities and the virulence of the whooping cough agent Bordetella pertussis. The RTX domain (RD) contains five RTX blocks (RTX-i to RTX-v) and their folding is driven by the binding of calcium. However, the detailed molecular mechanism via which the folding signal transmits within the five RTX blocks remains unknown. By combining single molecule optical tweezers, protein engineering, and toxin activity assays, here we demonstrate that the folding of the RD follows a strict hierarchy, with the folding starting from its C-terminal block RTX-v and proceeding towards the N-terminal RTX-i block sequentially. Our results reveal a strict series, templated folding mechanism, where the folding signal is transmitted along the RD in a series fashion from its C terminus continuously to the N terminus. Due to the series nature of this folding signal transmission pathway, the folding of RD can be disrupted at any given RTX block, rendering the RTX blocks located N-terminally to the disruption site and the acylation region of CyaA unfolded and abolishing CyaA's toxin activities. Our results reveal key mechanistic insights into the secretion and folding process of CyaA and may open up new potential avenues towards designing new therapeutics to abolish toxin activity of CyaA and combat B. pertussis.
Keywords: adenylate cyclase; bacterial toxin; optical tweezers; protein folding; single-molecule biophysics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interests with the contents of this article.
Figures




References
-
- Anderson E.L. Prevention of pertussis. Semin. Respir. Infect. 1989;4:284–292. - PubMed
-
- Frumkin K. Pertussis and persistent cough: practical, clinical and epidemiologic issues. J. Emerg. Med. 2013;44:889–895. - PubMed
-
- Weiss A.A., Hewlett E.L. Virulence factors of Bordetella pertussis. Annu. Rev. Microbiol. 1986;40:661–686. - PubMed
-
- Ladant D., Ullmann A. Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 1999;7:172–176. - PubMed
-
- Vojtova J., Kamanova J., Sebo P. Bordetella adenylate cyclase toxin: a swift saboteur of host defense. Curr. Opin. Microbiol. 2006;9:69–75. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

