FBXL4 mutations cause excessive mitophagy via BNIP3/BNIP3L accumulation leading to mitochondrial DNA depletion syndrome
- PMID: 37568009
- PMCID: PMC10589232
- DOI: 10.1038/s41418-023-01205-1
FBXL4 mutations cause excessive mitophagy via BNIP3/BNIP3L accumulation leading to mitochondrial DNA depletion syndrome
Abstract
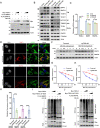
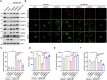
Mitochondria are essential organelles found in eukaryotic cells that play a crucial role in ATP production through oxidative phosphorylation (OXPHOS). Mitochondrial DNA depletion syndrome (MTDPS) is a group of genetic disorders characterized by the reduction of mtDNA copy number, leading to deficiencies in OXPHOS and mitochondrial functions. Mutations in FBXL4, a substrate-binding adaptor of Cullin 1-RING ubiquitin ligase complex (CRL1), are associated with MTDPS, type 13 (MTDPS13). Here, we demonstrate that, FBXL4 directly interacts with the mitophagy cargo receptors BNIP3 and BNIP3L, promoting their degradation through the ubiquitin-proteasome pathway via the assembly of an active CRL1FBXL4 complex. However, MTDPS13-associated FBXL4 mutations impair the assembly of an active CRL1FBXL4 complex. This results in a notable accumulation of BNIP3/3L proteins and robust mitophagy even at basal levels. Excessive mitophagy was observed in Knockin (KI) mice carrying a patient-derived FBXL4 mutation and cortical neurons (CNs)-induced from MTDPS13 patient human induced pluripotent stem cells (hiPSCs). In summary, our findings suggest that abnormal activation of BNIP3/BNIP3L-dependent mitophagy impairs mitochondrial homeostasis and underlies FBXL4-mutated MTDPS13.
© 2023. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- No. 91957125, 81972396/National Natural Science Foundation of China (National Science Foundation of China)
- 82272992 and 91954106/National Natural Science Foundation of China (National Science Foundation of China)
- 31821002, 31930062/National Natural Science Foundation of China (National Science Foundation of China)
- 82071269/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources

