Metabolites and Immune Response in Tumor Microenvironments
- PMID: 37568713
- PMCID: PMC10417674
- DOI: 10.3390/cancers15153898
Metabolites and Immune Response in Tumor Microenvironments
Abstract
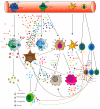
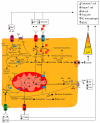
The remodeled cancer cell metabolism affects the tumor microenvironment and promotes an immunosuppressive state by changing the levels of macro- and micronutrients and by releasing hormones and cytokines that recruit immunosuppressive immune cells. Novel dietary interventions such as amino acid restriction and periodic fasting mimicking diets can prevent or dampen the formation of an immunosuppressive microenvironment by acting systemically on the release of hormones and growth factors, inhibiting the release of proinflammatory cytokines, and remodeling the tumor vasculature and extracellular matrix. Here, we discuss the latest research on the effects of these therapeutic interventions on immunometabolism and tumor immune response and future scenarios pertaining to how dietary interventions could contribute to cancer therapy.
Keywords: diets; immune system; metabolism.
Conflict of interest statement
V.D.L. holds intellectual property rights on clinical uses of FMD and equity interest in L-Nutra, a company that markets medical food.
Figures


References
-
- Mayers J.R., Torrence M.E., Danai L.V., Papagiannakopoulos T., Davidson S.M., Bauer M.R., Lau A.N., Ji B.W., Dixit P.D., Hosios A.M., et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. - DOI - PMC - PubMed
-
- Yuneva M.O., Fan T.W., Allen T.D., Higashi R.M., Ferraris D.V., Tsukamoto T., Mates J.M., Alonso F.J., Wang C., Seo Y., et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

