Plasma Exosome Proteins ILK1 and CD14 Correlated with Organ-Specific Metastasis in Advanced Gastric Cancer Patients
- PMID: 37568802
- PMCID: PMC10417498
- DOI: 10.3390/cancers15153986
Plasma Exosome Proteins ILK1 and CD14 Correlated with Organ-Specific Metastasis in Advanced Gastric Cancer Patients
Abstract
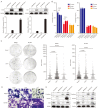
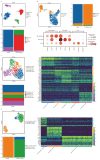
The exosome plays important roles in driving tumor metastasis, while the role of exosome proteins during organ-specific metastasis in gastric cancer has not been fully understood. To address this question, peripheral blood samples from 12 AGC patients with organ-specific metastasis, including distant lymphatic, hepatic and peritoneal metastasis, were collected to purify exosomes and to detect exosome proteins by Nano-HPLC-MS/MS. Gastric cancer cell lines were used for in vitro experiments. Peripheral blood sample and ascites sample from one patient were further analyzed by single-cell RNA sequencing. GO and KEGG enrichment analysis showed different expression proteins of hepatic metastasis were correlated with lipid metabolism. For peritoneal metastasis, actin cytoskeleton regulation and glycolysis/gluconeogenesis could be enriched. ILK1 and CD14 were correlated with hepatic and peritoneal metastasis, respectively. Overexpression of CD14 and ILK1 impacted the colony formation ability of gastric cancer and increased expression of Vimentin. CD14 derived from immune cells in malignant ascites correlated with high activation of chemokine- and cytokine-mediated signaling pathways. In summary, biological functions of plasma exosome proteins among AGC patients with different metastatic modes were distinct, in which ILK1 and CD14 were correlated with organ-specific metastasis.
Keywords: exosome; gastric cancer; organ-specific metastasis; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

