The N-Terminal Region of the BcWCL1 Photoreceptor Is Necessary for Self-Dimerization and Transcriptional Activation upon Light Stimulation in Yeast
- PMID: 37569251
- PMCID: PMC10418492
- DOI: 10.3390/ijms241511874
The N-Terminal Region of the BcWCL1 Photoreceptor Is Necessary for Self-Dimerization and Transcriptional Activation upon Light Stimulation in Yeast
Abstract
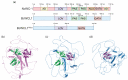
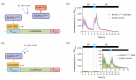
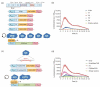
The BcWCL1 protein is a blue-light photoreceptor from the fungus Botrytis cinerea. This protein has a central role in B. cinerea circadian regulation and is an ortholog to WC-1 from Neurospora crassa. The BcWCL1 and WC-1 proteins have similar protein domains, including a LOV (Light Oxygen Voltage) domain for light sensing, two PAS (Per Arnt Sim) domains for protein-protein interaction, and a DNA binding domain from the GATA family. Recently, the blue-light response of BcWCL1 was demonstrated in a version without PAS domains (BcWCL1PAS∆). Here, we demonstrated that BcWCL1PAS∆ is capable of self-dimerization through its N-terminal region upon blue-light stimulation. Interestingly, we observed that BcWCL1PAS∆ enables transcriptional activation as a single component in yeast. By using chimeric transcription factors and the luciferase reporter gene, we assessed the transcriptional activity of different fragments of the N-terminal and C-terminal regions of BcWCL1PAS∆, identifying a functional transcriptional activation domain (AD) in the N-terminal region that belongs to the 9aaTAD family. Finally, we determined that the transcriptional activation levels of BcWCL1PAS∆ AD are comparable to those obtained with commonly used ADs in eukaryotic cells (Gal4 and p65). In conclusion, the BcWCL1PAS∆ protein self-dimerized and activated transcription in a blue-light-dependent fashion, opening future applications of this photoreceptor in yeast optogenetics.
Keywords: Botrytis cinerea; blue light; photoreceptor; transcriptional activation; yeast.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Interactions between Core Elements of the Botrytis cinerea Circadian Clock Are Modulated by Light and Different Protein Domains.J Fungi (Basel). 2022 May 6;8(5):486. doi: 10.3390/jof8050486. J Fungi (Basel). 2022. PMID: 35628742 Free PMC article.
-
Genome-Wide Characterization of Light-Regulated Gene Expression in Botrytis cinerea Reveals Underlying Complex Photobiology.Int J Mol Sci. 2023 May 13;24(10):8705. doi: 10.3390/ijms24108705. Int J Mol Sci. 2023. PMID: 37240051 Free PMC article.
-
Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins.Mol Microbiol. 1998 Aug;29(3):719-29. doi: 10.1046/j.1365-2958.1998.00955.x. Mol Microbiol. 1998. PMID: 9723912
-
[Photoreceptor apparatus of a fungus Neurospora crassa].Mol Biol (Mosk). 2005 Jul-Aug;39(4):602-17. Mol Biol (Mosk). 2005. PMID: 16083009 Review. Russian.
-
Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea.Fungal Genet Biol. 2010 Nov;47(11):917-21. doi: 10.1016/j.fgb.2010.05.003. Epub 2010 May 13. Fungal Genet Biol. 2010. PMID: 20471485 Review.
Cited by
-
Optogenetic Modification of Glycerol Production in Wine Yeast.ACS Synth Biol. 2025 Mar 21;14(3):719-728. doi: 10.1021/acssynbio.4c00654. Epub 2025 Feb 14. ACS Synth Biol. 2025. PMID: 39951366
References
-
- Suzuki Y., Oda Y. Inhibitory Loci of both Blue and near Ultraviolet Lights on Lateral-type Sclerotial Development in Botrytis cinerea. Jpn. J. Phytopathol. 1979;45:54–61. doi: 10.3186/jjphytopath.45.54. - DOI
-
- Tan K.K. Complete reversibility of sporulation by near ultraviolet and blue light in Botrytis cinerea. Trans. Br. Mycol. Soc. 1974;63:203–205. doi: 10.1016/S0007-1536(74)80159-2. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

