Transcriptional Regulators Controlling Virulence in Pseudomonas aeruginosa
- PMID: 37569271
- PMCID: PMC10418997
- DOI: 10.3390/ijms241511895
Transcriptional Regulators Controlling Virulence in Pseudomonas aeruginosa
Abstract
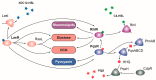
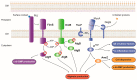
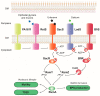
Pseudomonas aeruginosa is a pathogen capable of colonizing virtually every human tissue. The host colonization competence and versatility of this pathogen are powered by a wide array of virulence factors necessary in different steps of the infection process. This includes factors involved in bacterial motility and attachment, biofilm formation, the production and secretion of extracellular invasive enzymes and exotoxins, the production of toxic secondary metabolites, and the acquisition of iron. Expression of these virulence factors during infection is tightly regulated, which allows their production only when they are needed. This process optimizes host colonization and virulence. In this work, we review the intricate network of transcriptional regulators that control the expression of virulence factors in P. aeruginosa, including one- and two-component systems and σ factors. Because inhibition of virulence holds promise as a target for new antimicrobials, blocking the regulators that trigger the production of virulence determinants in P. aeruginosa is a promising strategy to fight this clinically relevant pathogen.
Keywords: Pseudomonas aeruginosa; one-component system; pathogenesis; quorum sensing; sigma factor; transcription regulators; two-component system; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

