SARS-CoV-2 Spike Protein Interaction Space
- PMID: 37569436
- PMCID: PMC10418891
- DOI: 10.3390/ijms241512058
SARS-CoV-2 Spike Protein Interaction Space
Abstract
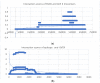
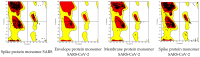
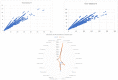
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a +sense single-strand RNA virus. The virus has four major surface proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N), respectively. The constitutive proteins present a high grade of symmetry. Identifying a binding site is difficult. The virion is approximately 50-200 nm in diameter. Angiotensin-converting enzyme 2 (ACE2) acts as the cell receptor for the virus. SARS-CoV-2 has an increased affinity to human ACE2 compared with the original SAR strain. Topological space, and its symmetry, is a critical component in molecular interactions. By exploring this space, a suitable ligand space can be characterized accordingly. A spike protein (S) computational model in a complex with ACE 2 was generated using silica methods. Topological spaces were probed using high computational throughput screening techniques to identify and characterize the topological space of both SARS and SARS-CoV-2 spike protein and its ligand space. In order to identify the symmetry clusters, computational analysis techniques, together with statistical analysis, were utilized. The computations are based on crystallographic protein data bank PDB-based models of constitutive proteins. Cartesian coordinates of component atoms and some cluster maps were generated and analyzed. Dihedral angles were used in order to compute a topological receptor space. This computational study uses a multimodal representation of spike protein interactions with some fragment proteins. The chemical space of the receptors (a dimensional volume) suggests the relevance of the receptor as a drug target. The spike protein S of SARS and SARS-CoV-2 is analyzed and compared. The results suggest a mirror symmetry of SARS and SARS-CoV-2 spike proteins. The results show thatSARS-CoV-2 space is variable and has a distinct topology. In conclusion, surface proteins grant virion variability and symmetry in interactions with a potential complementary target (protein, antibody, ligand). The mirror symmetry of dihedral angle clusters determines a high specificity of the receptor space.
Keywords: COVID-19; QSAR; SARS-CoV-2; antibody; antibody binding; chemical space; paratope; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
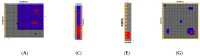
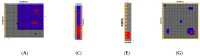

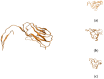




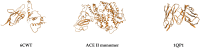
Similar articles
-
Hot spot profiles of SARS-CoV-2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method.Sci Rep. 2020 Oct 8;10(1):16862. doi: 10.1038/s41598-020-73820-8. Sci Rep. 2020. PMID: 33033344 Free PMC article.
-
Coevolution, Dynamics and Allostery Conspire in Shaping Cooperative Binding and Signal Transmission of the SARS-CoV-2 Spike Protein with Human Angiotensin-Converting Enzyme 2.Int J Mol Sci. 2020 Nov 4;21(21):8268. doi: 10.3390/ijms21218268. Int J Mol Sci. 2020. PMID: 33158276 Free PMC article.
-
Computational Alanine Scanning and Structural Analysis of the SARS-CoV-2 Spike Protein/Angiotensin-Converting Enzyme 2 Complex.ACS Nano. 2020 Sep 22;14(9):11821-11830. doi: 10.1021/acsnano.0c04674. Epub 2020 Aug 26. ACS Nano. 2020. PMID: 32833435 Free PMC article.
-
Potential therapeutic approaches for the early entry of SARS-CoV-2 by interrupting the interaction between the spike protein on SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2).Biochem Pharmacol. 2021 Oct;192:114724. doi: 10.1016/j.bcp.2021.114724. Epub 2021 Aug 8. Biochem Pharmacol. 2021. PMID: 34371003 Free PMC article. Review.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

