Molecular Modeling Unveils the Effective Interaction of B-RAF Inhibitors with Rare B-RAF Insertion Variants
- PMID: 37569660
- PMCID: PMC10418914
- DOI: 10.3390/ijms241512285
Molecular Modeling Unveils the Effective Interaction of B-RAF Inhibitors with Rare B-RAF Insertion Variants
Abstract
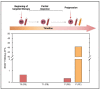
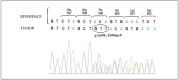
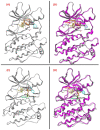
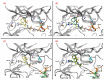
The Food and Drug Administration (FDA) has approved MAPK inhibitors as a treatment for melanoma patients carrying a mutation in codon V600 of the BRAF gene exclusively. However, BRAF mutations outside the V600 codon may occur in a small percentage of melanomas. Although these rare variants may cause B-RAF activation, their predictive response to B-RAF inhibitor treatments is still poorly understood. We exploited an integrated approach for mutation detection, tumor evolution tracking, and assessment of response to treatment in a metastatic melanoma patient carrying the rare p.T599dup B-RAF mutation. He was addressed to Dabrafenib/Trametinib targeted therapy, showing an initial dramatic response. In parallel, in-silico ligand-based homology modeling was set up and performed on this and an additional B-RAF rare variant (p.A598_T599insV) to unveil and justify the success of the B-RAF inhibitory activity of Dabrafenib, showing that it could adeptly bind both these variants in a similar manner to how it binds and inhibits the V600E mutant. These findings open up the possibility of broadening the spectrum of BRAF inhibitor-sensitive mutations beyond mutations at codon V600, suggesting that B-RAF V600 WT melanomas should undergo more specific investigations before ruling out the possibility of targeted therapy.
Keywords: BRAF rare mutations; advanced melanoma; ligand-based homology modeling; liquid biopsy; molecular docking calculation; targeted therapy.
Conflict of interest statement
M.C.S. received a travel grant from Agilent; all the other authors declare no conflicts of interest. This work is not in any way sponsored or to any extent influenced by any of the authors’ employment statuses.
Figures

References
-
- Johnson D.B., Zhao F., Noel M., Riely G.J., Mitchell E.P., Wright J.J., Chen H.X., Gray R.J., Li S., McShane L.M., et al. Trametinib Activity in Patients with Solid Tumors and Lymphomas Harboring BRAF Non-V600 Mutations or Fusions: Results from NCI-MATCH (EAY131) Clin. Cancer Res. 2020;26:1812–1819. doi: 10.1158/1078-0432.CCR-19-3443. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

