A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells
- PMID: 37569739
- PMCID: PMC10418425
- DOI: 10.3390/ijms241512364
A Sphingolipidomic Profiling Approach for Comparing X-ray-Exposed and Unexposed HepG2 Cells
Abstract
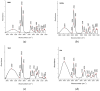
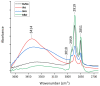
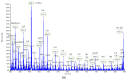
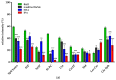
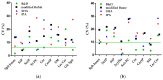
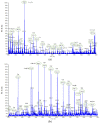
An analytical method based on tandem mass spectrometry-shotgun is presently proposed to obtain sphingolipidomic profiles useful for the characterization of lipid extract from X-ray-exposed and unexposed hepatocellular carcinoma cells (HepG2). To obtain a targeted lipidic profile from a specific biological system, the best extraction method must be identified before instrumental analysis. Accordingly, four different classic lipid extraction protocols were compared in terms of efficiency, specificity, and reproducibility. The performance of each procedure was evaluated using the Fourier-transform infrared spectroscopic technique; subsequently, the quality of extracts was estimated using electrospray ionization tandem mass spectrometry. The selected procedure based on chloroform/methanol/water was successfully used in mass spectrometry-based shotgun sphingolipidomics, allowing for evaluation of the response of cells to X-ray irradiation, the most common anticancer therapy. Using a relative quantitative approach, the changes in the sphingolipid profiles of irradiated cell extracts were demonstrated, confirming that lipidomic technologies are also useful tools for studying the key sphingolipid role in regulating cancer growth during radiotherapy.
Keywords: HepG2 cells; X-ray; extraction methods; mass spectrometry; sphingolipidomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Custodia A., Romaus-Sanjurjo D., Aramburu-Núñez M., Álvarez-Rafael D., Vázquez-Vázquez L., Camino-Castiñeiras J., Leira Y., Pías-Peleteiro J.M., Aldrey J.M., Sobrino T., et al. Ceramide/Sphingosine 1-Phosphate Axis as a Key Target for Diagnosis and Treatment in Alzheimer’s Disease and Other Neurodegenerative Diseases. Int. J. Mol. Sci. 2022;23:8082. doi: 10.3390/ijms23158082. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

