Generating Potential Protein-Protein Interaction Inhibitor Molecules Based on Physicochemical Properties
- PMID: 37570623
- PMCID: PMC10420264
- DOI: 10.3390/molecules28155652
Generating Potential Protein-Protein Interaction Inhibitor Molecules Based on Physicochemical Properties
Abstract
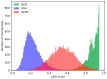
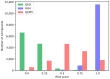
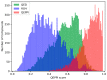
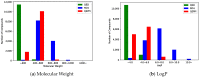
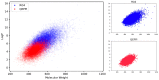
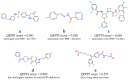
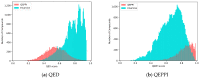
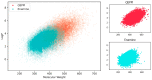
Protein-protein interactions (PPIs) are associated with various diseases; hence, they are important targets in drug discovery. However, the physicochemical empirical properties of PPI-targeted drugs are distinct from those of conventional small molecule oral pharmaceuticals, which adhere to the "rule of five (RO5)". Therefore, developing PPI-targeted drugs using conventional methods, such as molecular generation models, is challenging. In this study, we propose a molecular generation model based on deep reinforcement learning that is specialized for the production of PPI inhibitors. By introducing a scoring function that can represent the properties of PPI inhibitors, we successfully generated potential PPI inhibitor compounds. These newly constructed virtual compounds possess the desired properties for PPI inhibitors, and they show similarity to commercially available PPI libraries. The virtual compounds are freely available as a virtual library.
Keywords: QEPPI; molecular generation; protein-protein interaction inhibitor; rule of five; rule of four; virtual chemical library.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

