Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells
- PMID: 37570631
- PMCID: PMC10419799
- DOI: 10.3390/molecules28155658
Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells
Abstract
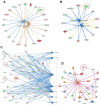
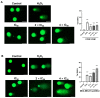
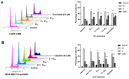
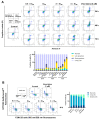
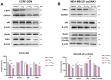
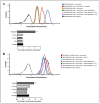
The c-MYC oncogene regulates multiple cellular activities and is a potent driver of many highly aggressive human cancers, such as leukemia and triple-negative breast cancer. The oxadiazole class of compounds has gained increasing interest for its anticancer activities. The aim of this study was to investigate the molecular modes of action of a 1,2,4-oxadiazole derivative (ZINC15675948) as a c-MYC inhibitor. ZINC15675948 displayed profound cytotoxicity at the nanomolar range in CCRF-CEM leukemia and MDA-MB-231-pcDNA3 breast cancer cells. Multidrug-resistant sublines thereof (i.e., CEM/ADR5000 and MDA-MB-231-BCRP) were moderately cross-resistant to this compound (<10-fold). Molecular docking and microscale thermophoresis revealed a strong binding of ZINC15675948 to c-MYC by interacting close to the c-MYC/MAX interface. A c-MYC reporter assay demonstrated that ZINC15675948 inhibited c-MYC activity. Western blotting and qRT-PCR showed that c-MYC expression was downregulated by ZINC15675948. Applying microarray hybridization and signaling pathway analyses, ZINC15675948 affected signaling routes downstream of c-MYC in both leukemia and breast cancer cells as demonstrated by the induction of DNA damage using single cell gel electrophoresis (alkaline comet assay) and induction of apoptosis using flow cytometry. ZINC15675948 also caused G2/M phase and S phase arrest in CCRF-CEM cells and MDA-MB-231-pcDNA3 cells, respectively, accompanied by the downregulation of CDK1 and p-CDK2 expression using western blotting. Autophagy induction was observed in CCRF-CEM cells but not MDA-MB-231-pcDNA3 cells. Furthermore, microarray-based mRNA expression profiling indicated that ZINC15675948 may target c-MYC-regulated ubiquitination, since the novel ubiquitin ligase (ELL2) was upregulated in the absence of c-MYC expression. We propose that ZINC15675948 is a promising natural product-derived compound targeting c-MYC in c-MYC-driven cancers through DNA damage, cell cycle arrest, and apoptosis.
Keywords: 1,2,4-oxadiazole; c-MYC inhibitor; leukemia; natural product derivative; oncogenes; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Sammak S., Hamdani N., Gorrec F., Allen M.D., Freund S.M.V., Bycroft M., Zinzalla G. Crystal structures and nuclear magnetic resonance studies of the apo form of the c-MYC: MAX bHLHZip complex reveal a helical basic region in the absence of DNA. Biochemistry. 2019;58:3144–3154. doi: 10.1021/acs.biochem.9b00296. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

